Aptamer realizing specific binding with newcastle disease virus as well as screening method and applications of aptamer
A nucleic acid aptamer, Newcastle disease virus technology, applied in the field of molecular biology, can solve the problems affecting the efficiency of screening and ligand affinity, and achieve the effects of shortening incubation time, reducing differences, and improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1. In vitro screening of Newcastle disease virus nucleic acid aptamers
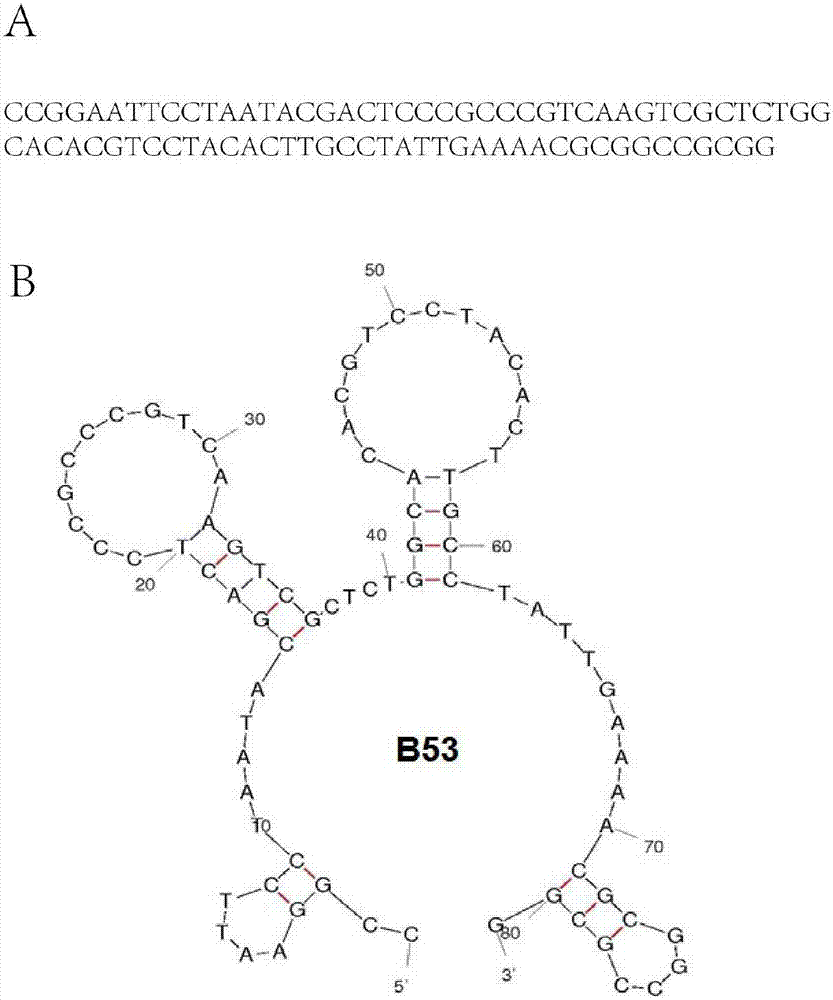
[0047] S1. Construct a random nucleic acid library with a synthetic length of 81 nt, the sequence of which is shown in SEQ ID NO.1, 5'-CCGGAATTCCTAATACGACTC–(N)40–TATTGAAAACGCGGCCGCGG-3'; where the two ends are 20 and 21 bases respectively The sequence is fixed, and the middle 40 bases are random sequences.
[0048] The sequences of the upstream primer F and the downstream primer R synthesized for PCR amplification are:
[0049] Upstream primer F: 5'-CCGGAATTCCTAATACGACTC-3', as shown in SEQ ID NO.2;
[0050] Downstream primer R: 5'-CCGCGGCCGCGTTTTCAATA-3', as shown in SEQ ID NO.3;
[0051] Biotin-modified upstream primer Biotin-F: 5'-Biotin-CCGGAATTCCTAATACGACTC-3', as shown in SEQ ID NO.4.
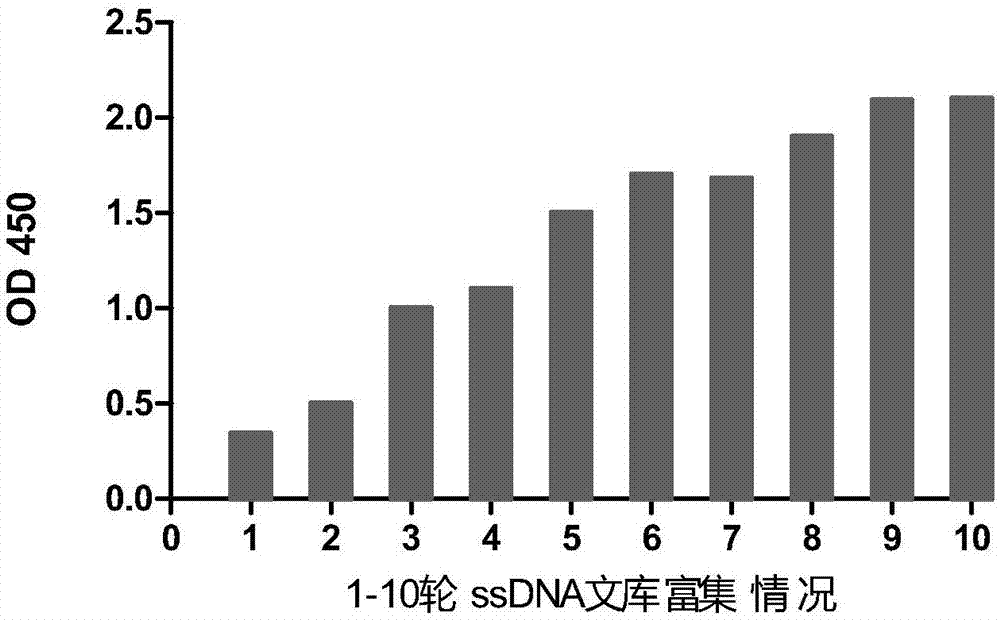
[0052] S2. Incubation and separation of ssDNA library and HN protein, add 35.5 μg ssDNA library to 100 μL binding buffer (50 mM Tris-HCl, 25 mM NaCl, 5 mM MgCl2, 10 mM DTT (dithiothreitol), pH 7.5)...
Embodiment 2
[0063] Example 2. Enzyme-linked immunosorbent assay (ELISA)
[0064] The nucleic acid aptamers obtained by screening were sent to Sangon Bioengineering (Shanghai) Co., Ltd. for synthesis, modified with biotin, and tested for specificity by ELISA method. Different samples were selected, the coating volume was 2 μL / well, and a blank control group was set up at the same time. Coat overnight at 4°C, block with 1% BSA at 37°C, add different biotin-labeled aptamers to the wells for incubation, then add horseradish peroxidase-labeled avidin for incubation, add substrate The substance was used for color development, the color development was terminated, and the OD was measured with a microplate reader 450 value.
[0065] image 3 The ELISA detection of the aptamer B53 screened for SELEX, Figure A shows the affinity of the aptamer to different strains of Newcastle disease virus F48E9, GM and La Sota. like image 3 As shown, the results show that the aptamer B53 has affinity to New...
Embodiment 3
[0066] Embodiment 3. Dot-blot test (Dot-blot)
[0067] Spot 2 μL of HN protein (1mg / mL), GM strain purified virus, AIV, IBV, SPF chick embryo allantoic fluid onto the NC membrane, and set up a blank control group without dripping the solution; after the NC membrane is dry, Add 3% BSA blocking solution to block at 37°C for 2 hours, wash the membrane with PBST solution; add 5 pmol / μL biotin-labeled ssDNA to the NC membrane, incubate at 37°C for 2 hours, then wash the membrane with PBST solution ; Add the NC membrane to the 1:5000 diluted Streptavidin-HRP solution, incubate at 37°C for 1 hour, then wash the membrane with PBST solution for 3 minutes, each time for 3 minutes; stop the color development after 10 minutes of DAB display, take out the membrane, Observe the color development of the membrane.
[0068] The results of aptamer specificity analysis were as follows: Figure 4 as shown, Figure 4 A represents the results of the aptamer against different strains of Newcastle...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com