Method for detecting sheep FecB gene polymorphism by using micro-fluidic SNP chip
A gene polymorphism and chip detection technology, which is applied in the determination/testing of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problems of difficult high-throughput automatic determination, numerous procedures, and high cost. Achieve the effect of reducing manual operation process, easy interpretation and simplified operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044]Embodiment 1 is aimed at the design of detection primer and probe of sheep FecB gene SNP site
[0045] A single nucleotide polymorphism A / G was carried out for the nucleotide of the 29382188bp site (NC_019463.1, based on the sheep genome sequence information version number Oar_v3.1, December 2012) on the No. 6 chromosome of sheep, and the designed The following primers and probe combinations were used to detect the polymorphism of sheep FecB gene by microfluidic SNP chip.
[0046] A total of 5 pairs of primers and probes were designed based on the polymorphic site sequence of the FecB gene, and fluorescent reporter groups of different colors were labeled on the 5' of the probes.
[0047] The nucleotide sequence of the primer 1 is as follows:
[0048] Forward primer: 5'-CCAGCTGGTTCCGAGAGACA-3'
[0049] Reverse primer: 5'-CTTATACTCACCCAAGATGTTTTCATG-3'
[0050] The nucleotide sequence of the probe 1 is as follows:
[0051] P-G: 5'-FAM-AAATATATCGGACGGTGTT-MGB-3'
[005...
Embodiment 2
[0078] Example 2 Method for detecting sheep FecB gene polymorphism using microfluidic SNP chip
[0079] 1. Experimental materials A total of 95 sheep produced by crossbreeding Small-tailed Han sheep as the female parent and other breeds as the male parent were selected as the test objects.
[0080] 2. Extraction of Genomic DNA 1ml of sheep blood was collected from the jugular vein and anticoagulated with EDTA. First, the red blood cell lysate is lysed to remove DNA-free red blood cells. The nucleus lysate lyses the cells to release genomic DNA. Then the protein precipitation solution selectively precipitates to remove the protein. The pure genomic DNA is precipitated by isopropanol and redissolved in the DNA solution.
[0081] 3. Microfluidic SNP chip for genotyping
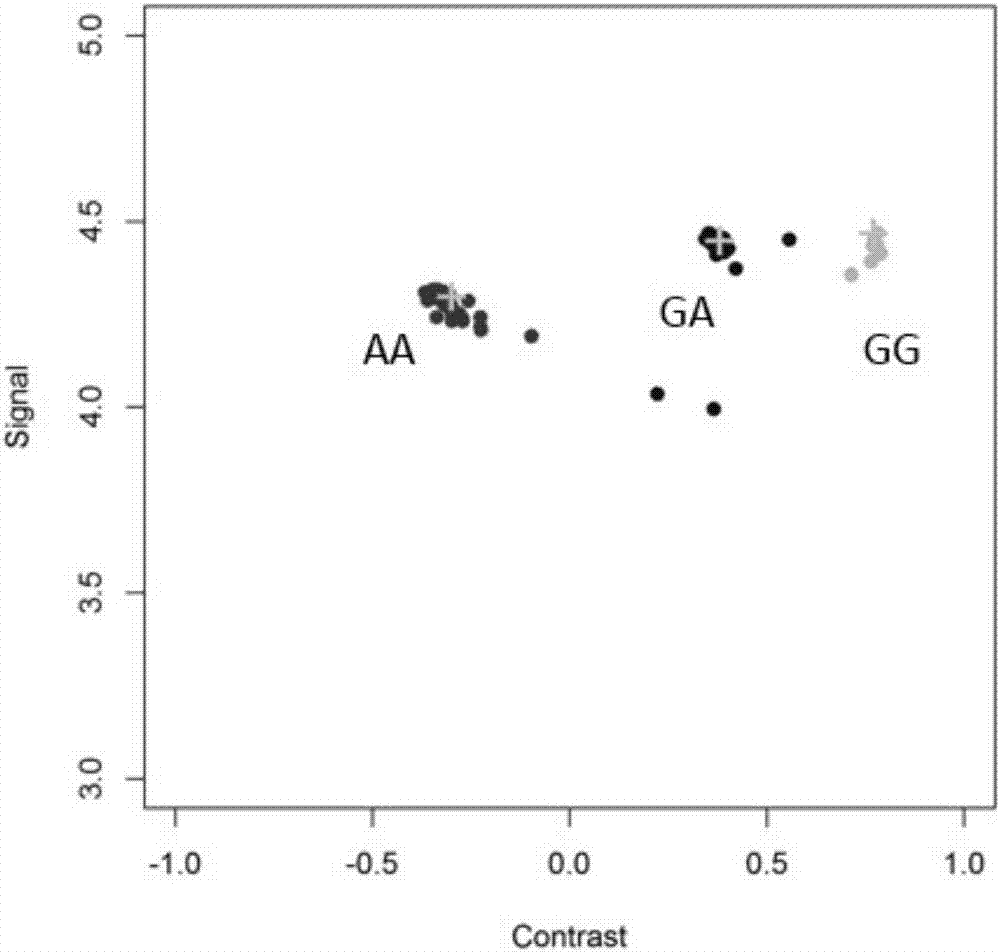
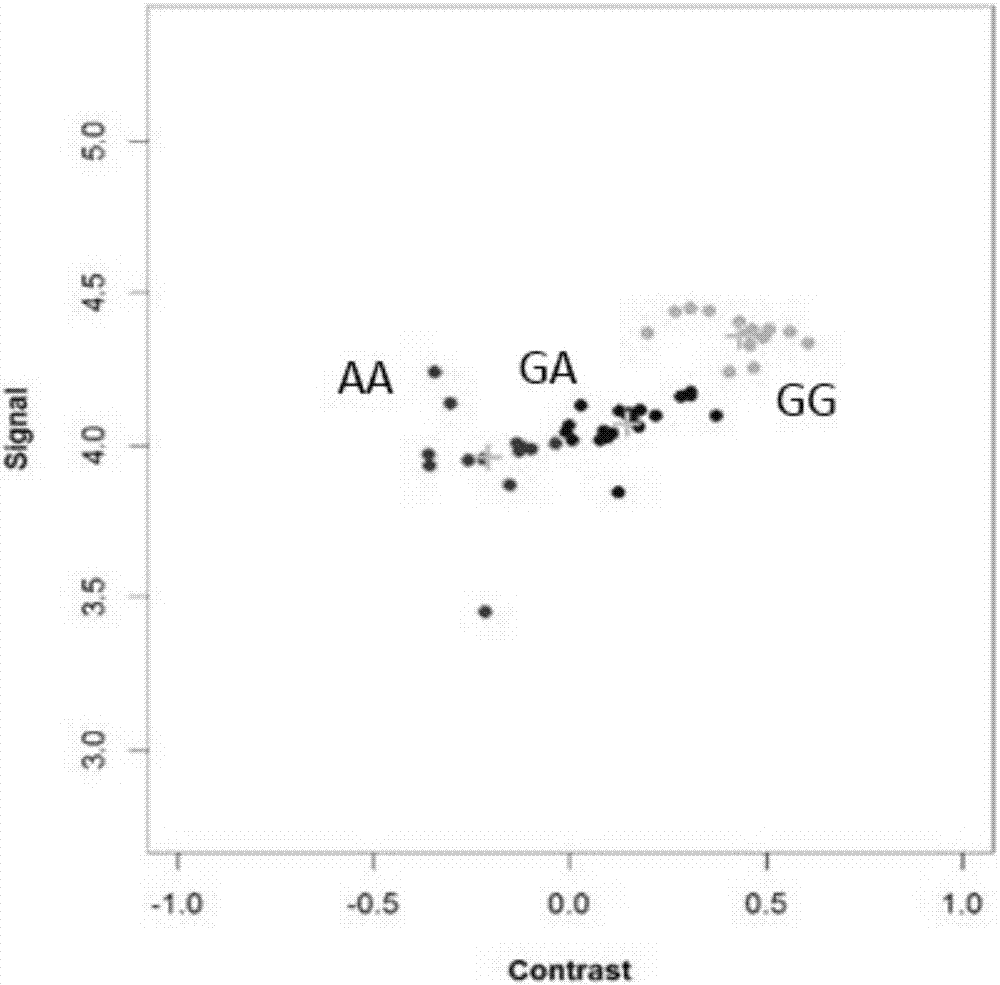
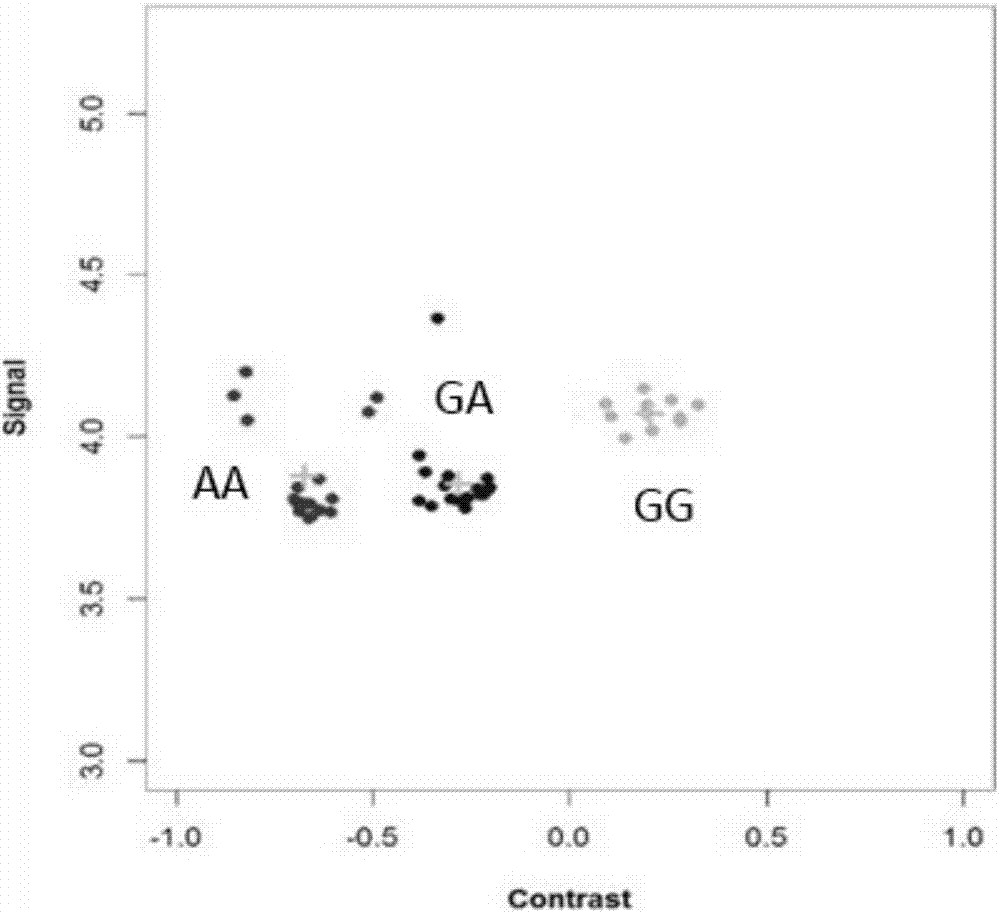
[0082] The best detection primers and probes for the 29382188bp site on sheep chromosome 6 (NC_019463.1, based on the sheep genome sequence information version number Oar_v3.1, December 2012) screened in Example...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com