Kit for extracting trace DNA and extraction method
A kit and trace technology, which is applied in the field of trace DNA detection, can solve the problems of DNA sample loss, complicated operation, and less number of transfer tubes, and achieve the effects of reducing trace DNA loss, improving extraction efficiency and slow sedimentation speed.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
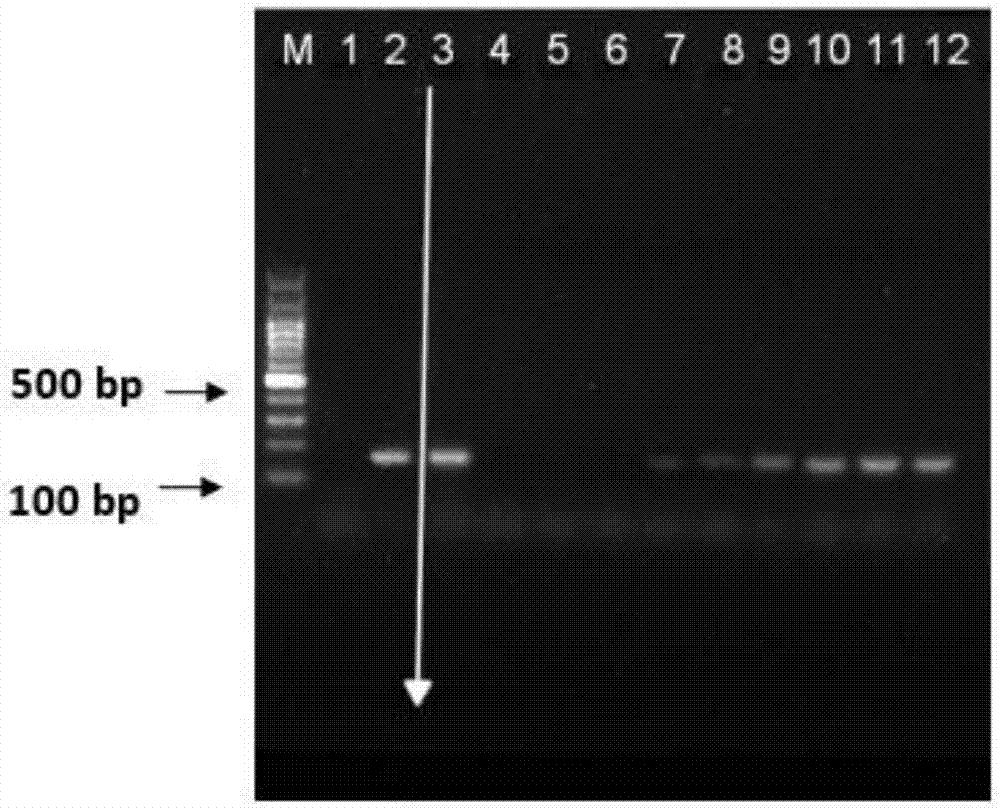
[0055] Example 1 Internal Control PCR System Verification of Minimum Loading Quantity of Trace DNA
[0056] Experiments were designed to verify the internal control PCR system with the lowest loading amount of trace DNA, and determine the minimum amount of template that can carry out effective PCR reactions.
[0057] Genomic DNA templates of 50ng / μl volunteers were serially diluted to obtain a total of 3000pg, 2000pg, 1000pg, 500pg, 100pg, 50pg, 30pg, 10pg, and 5pg of genomic DNA templates for verification. In order to ensure the accuracy of the experiment, a control experiment was designed: the negative control template was 10mmol / L Tris, which was used to check whether there was nucleic acid contamination in the reaction system; the positive control template was 50ng / μl human genomic DNA, which was purchased from Kangwei Century Human GenomicDNA is used to check whether the amplification ability of the reaction system is normal and whether the sample is abnormal; plus a tota...
Embodiment 2
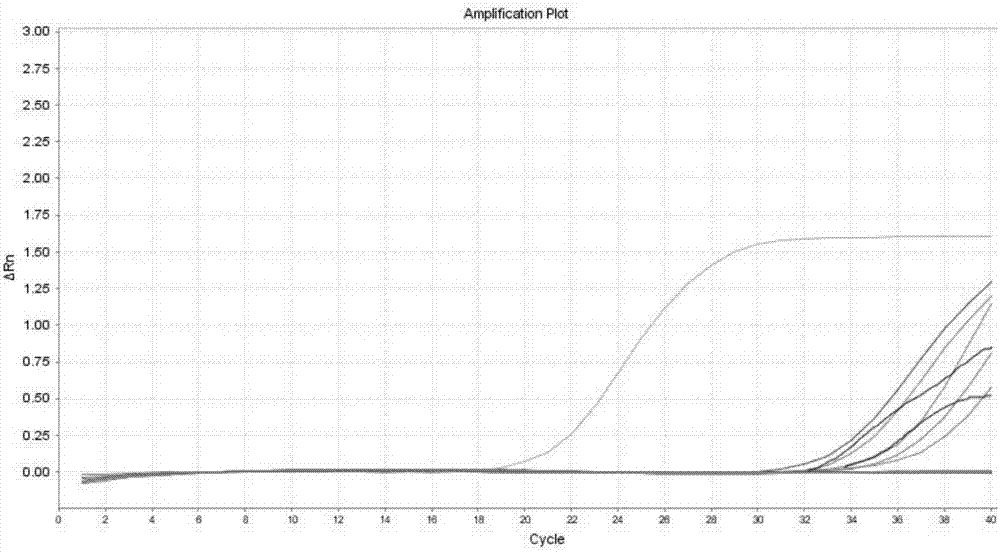
[0068] Embodiment 2 Reagent component concentration is verified to detection rate
[0069] Design an orthogonal experiment to determine the concentration of NaCl, guanidine isothiocyanate and sodium dodecylsulfonate in the lysate to verify the extraction rate of trace DNA, design an orthogonal experiment with 3 factors and 4 levels; Nacl 0.1mol / L , 0.5mol / L, 1mol / L, 2mol / L; isothiocyanate concentration: 2mol / L, 3mol / L, 4mol / L, 5mol / L; SDS concentration: 1%, 2%, 5%, 10%
[0070] In the experiment, 10^6 cells / ml of cultured cells were used, diluted 10 times, and about 100 cells were extracted from 1 μl of cell suspension
[0071] (1) Add a suspension containing 100 cultured cells to a 1.5ml centrifuge tube;
[0072] (2) To the centrifuge tube with the template added in (1), respectively add the lysate composed of NaCl, guanidine isothiocyanate and sodium dodecylsulfonate at different concentrations according to the orthogonal experiment table, add proteinase K, and pipette mix...
Embodiment 3
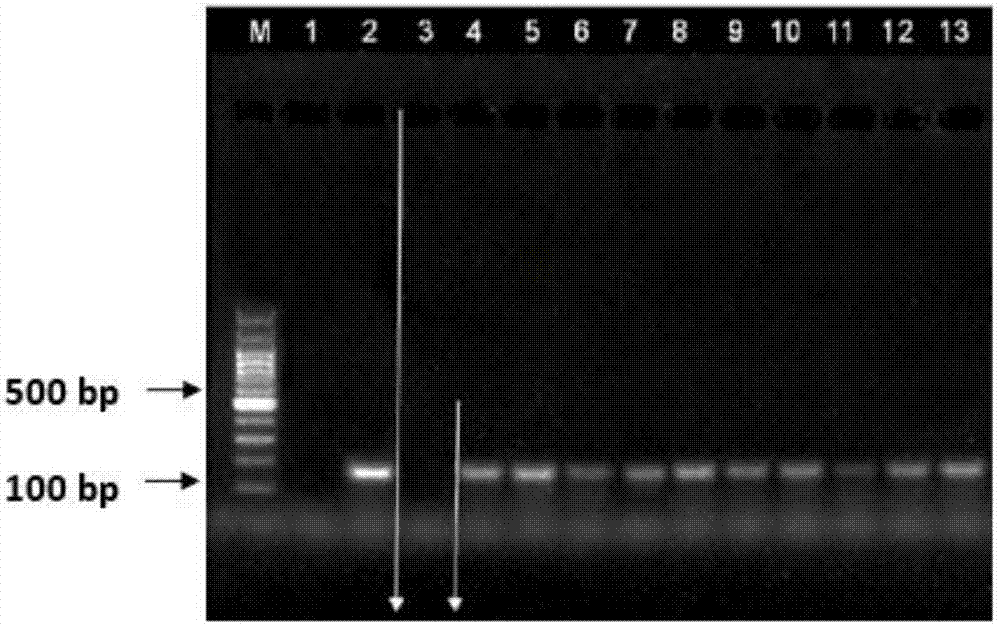
[0085] Example 3 micro-cell extraction verification
[0086] The experimental sample is the cultured lung cancer cell line NCI-H1975, the concentration of cultured cells is 10^6 cells / ml, diluted 100 times, and about 10 cells are taken from 1 μl of cell suspension for extraction.
[0087] (1) Add 1 μl suspension containing 10 cultured cells to a 1.5ml centrifuge tube;
[0088] (2) Add 250 μl of lysate and 10 μl of proteinase K;
[0089](3) Using the high-salt environment of the lysate, fully lyse the cells at 2000rpm, 55°C, and 20min to release the genomic DNA of the cells, then add isopropanol of the same volume as the lysate to the lysed mixture and pipette mix;
[0090] (4) Add 7 μl extraction magnetic beads and 10 μl nucleic acid sedimentation aid to the mixed solution obtained in the centrifuge tube, cover the centrifuge tube cap, place the centrifuge tube on a vertical mixer, and mix at room temperature for 10 minutes to allow the magnetic beads and genomic DNA fully ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com