Method for producing phloretin by fermentation of saccharomyces cerevisiae
A technology for Saccharomyces cerevisiae and Saccharomyces cerevisiae, which is applied in the field of Saccharomyces cerevisiae fermentation to produce phloretin, can solve problems such as serious pollution and serious chemical pollution, and achieve the effects of good purity, high content and simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1 Plasmid construction.
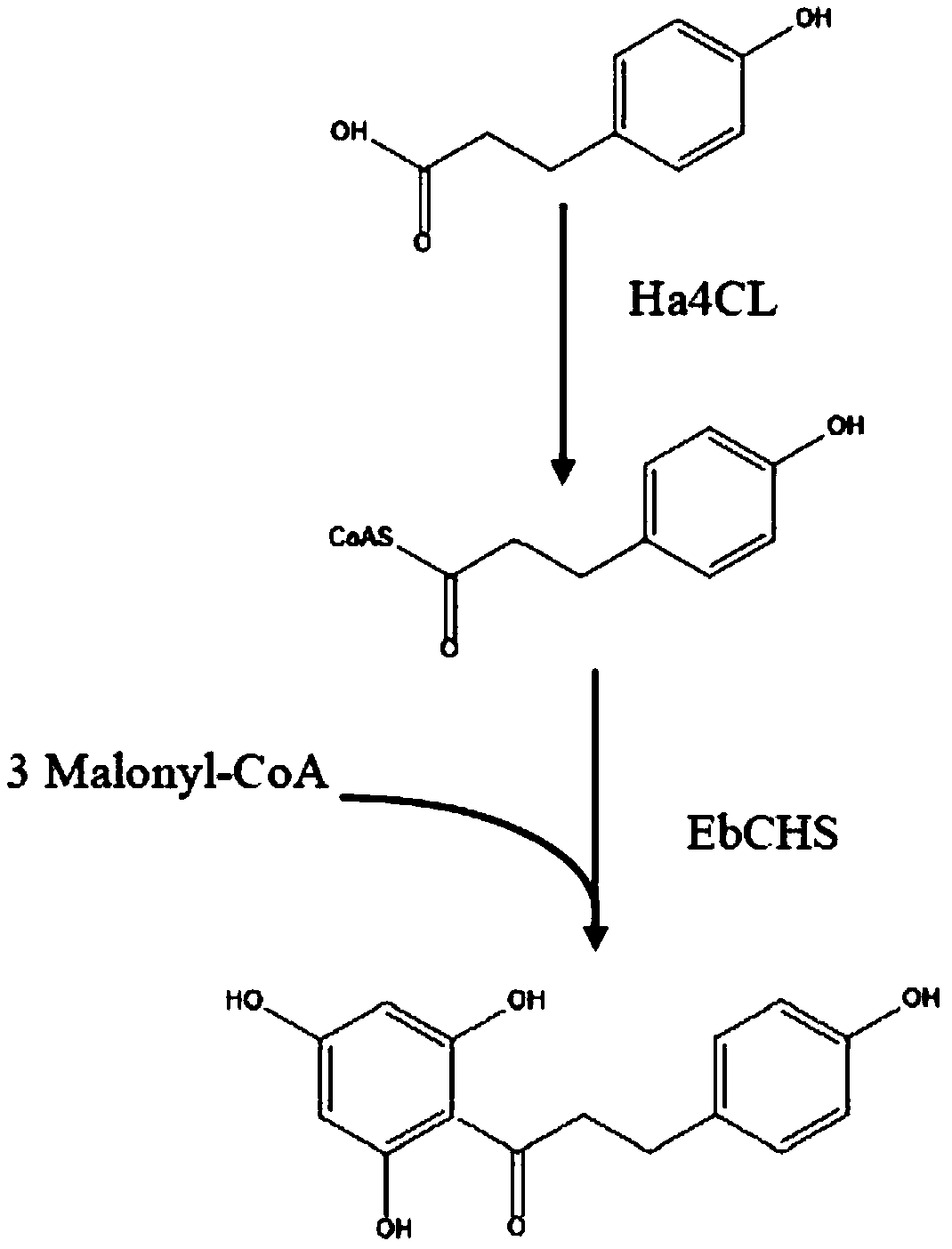
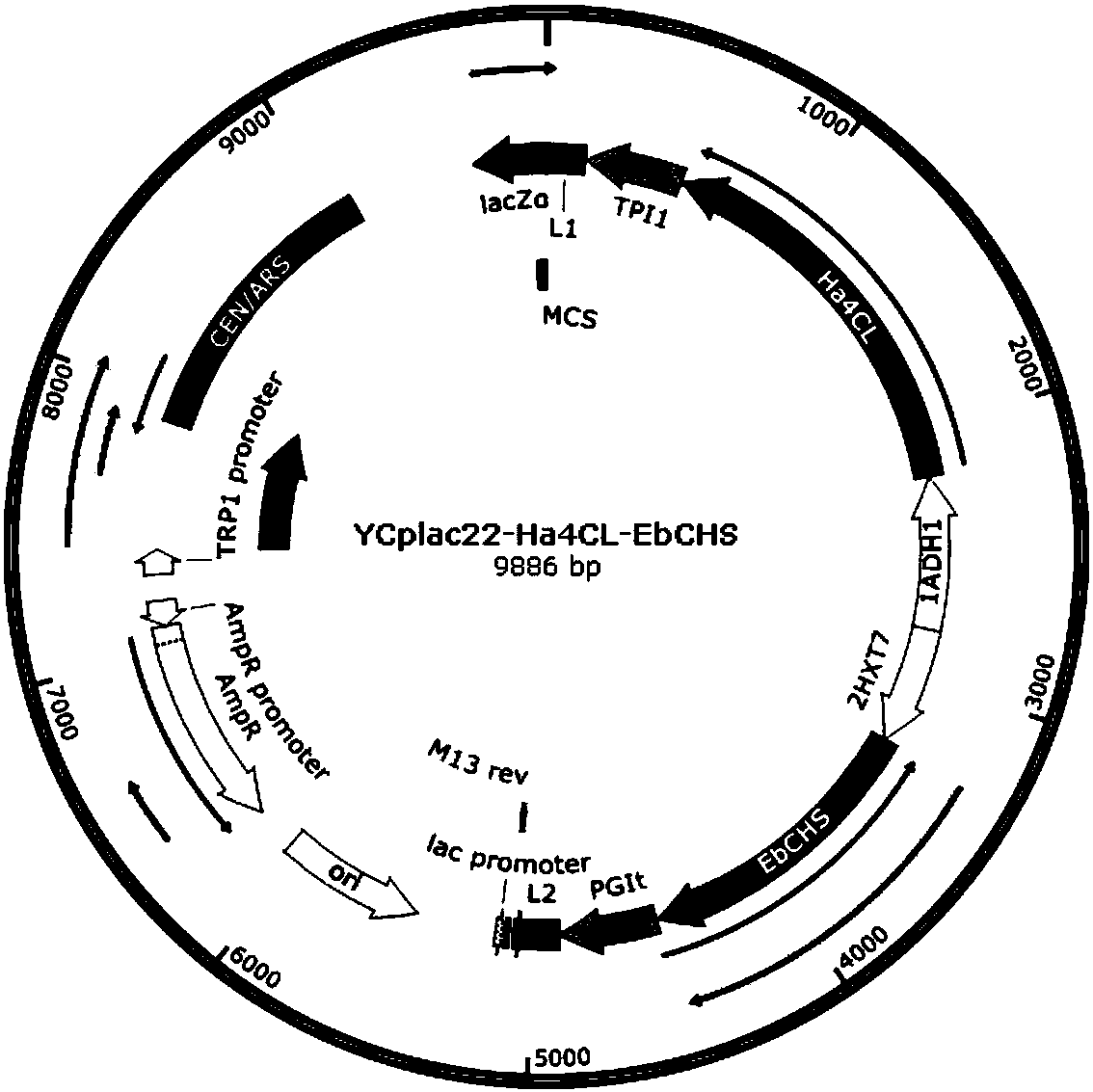
[0043] Design the nucleic acid sequence of the corresponding gene according to the amino acid sequence provided in the present invention and Saccharomyces cerevisiae codon preference, send the gene Ha4CL and EbCHS to a commercial company for synthesis, design a homology arm with a size of about 20bp, and use a one-step cloning kit to clone the two The gene is constructed on the vector YCplac22 to obtain vector 1, as shown in Figure 2 (a), the expression vector YCplac22 has the ampicillin resistance of Escherichia coli and the selection marker trp gene of Saccharomyces cerevisiae;
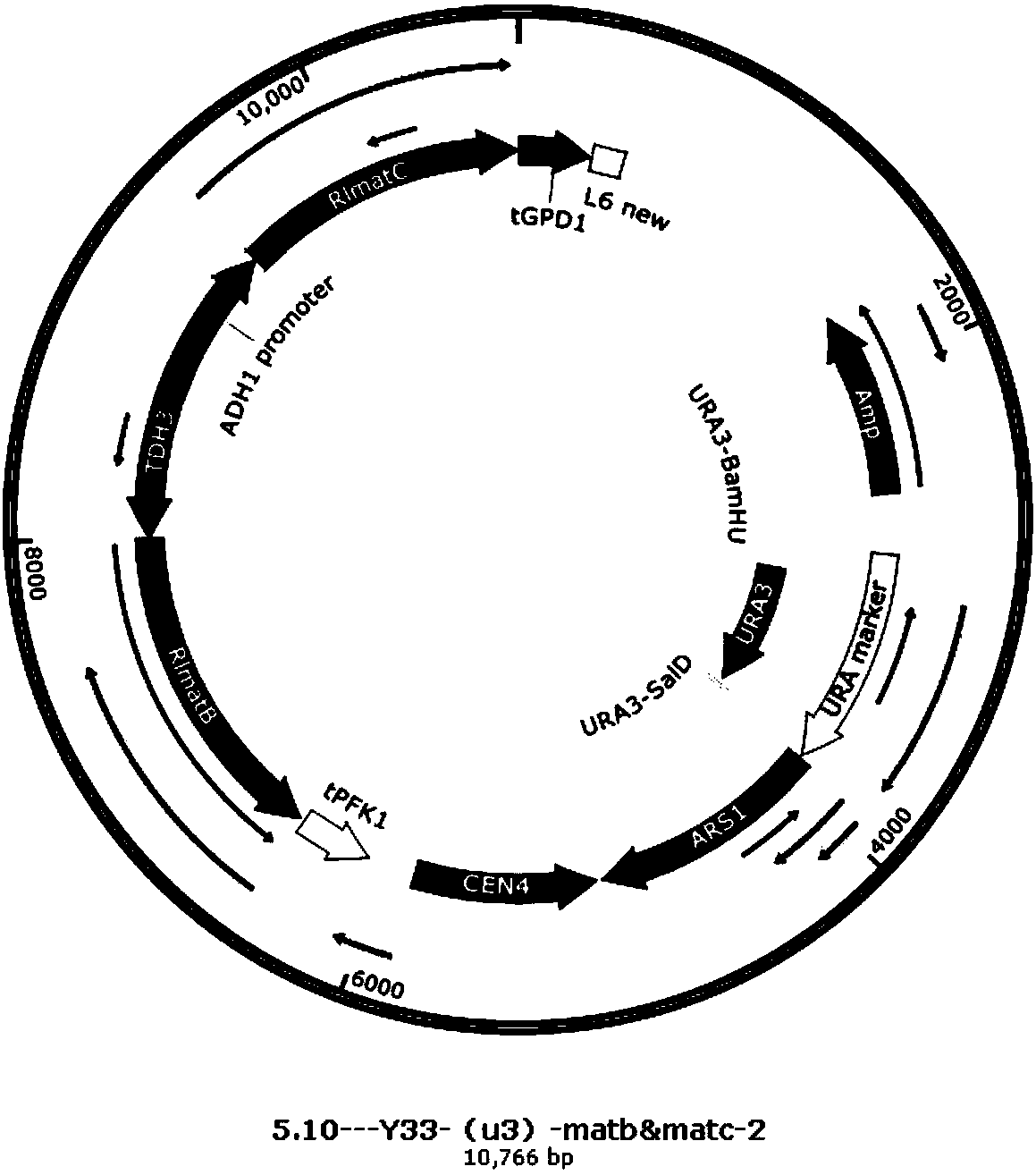
[0044] Two genes, RlmatB and RlmatC, which convert malonate from rhizobia of leguminous plants into malonyl-CoA, were optimized according to the codon preference of S. Obtain vector 2, as shown in Figure 2 (b), the expression vector YCplac33 has the ampicillin resistance gene of Escherichia coli and the selection marker ura gene of Saccharomyces cerevisiae; ...
Embodiment 2
[0046] Embodiment 2 conversion process.
[0047] 1) Pick a single colony and shake it overnight for 12 hours in the corresponding medium;
[0048] 2) measure the OD600 value of the bacterium liquid that cultivates with spectrophotometer;
[0049] 3) Transfer to 50 mL of fresh YPAD medium with an initial OD600 value of 0.2 OD;
[0050] 4) Activate for 4-5 hours so that the OD600 value of the two-generation bacterial liquid of Saccharomyces cerevisiae is 0.8-0.9;
[0051] Centrifuge at 3600rpm for 5min to collect the bacteria, 25mL ddH 2 O washed twice (ddH used 2 O is preferably sterilized on the same day);
[0052]5) Resuspend 1mL of water into a sterile 1.5mL centrifuge tube; (for use in ultra-clean bench)
[0053] 6) Centrifuge at 13000rpm for 30s to collect the bacteria;
[0054] 7) Resuspend in 1 mL of water and aliquot 100 μL per tube for transformation, centrifuge the aliquoted bacterial solution in a hand-held centrifuge for 20 seconds, discard the supernatant and...
Embodiment 3
[0065] Embodiment 3 integration process
[0066] 1) Saccharomyces cerevisiae genome was extracted, and the upstream and downstream homology arms of the YPRCdelta15 integration site on chromosome 16 of Saccharomyces cerevisiae were extracted by PCR to extract fragments, upstream fragment 1 and downstream fragment 2.
[0067] 2) The upstream fragment and the screening tag leu gene, gene (including promoter and terminator) SeACS L641P , ScADH2, ScALD6, ScACCl S659A,S1157A The homologous arm is transferred into Saccharomyces cerevisiae W303. In Saccharomyces cerevisiae, using homologous recombination, Saccharomyces cerevisiae itself will integrate the introduced gene fragment into the genome of Saccharomyces cerevisiae.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com