Enterobacter cloacae 16SrDNA and primer group Yg2 and application thereof in aspect of enterobacter cloacae molecule detection
A technology of Enterobacter cloacae and primer set, which is applied in the direction of recombinant DNA technology, DNA/RNA fragments, microbial determination/inspection, etc., can solve the problems of insufficient research technology, achieve high sensitivity, reliable detection results, and good sensitivity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1 Detection primer design and establishment of LAMP amplification method
[0053] 1. Primer design
[0054] In the present invention, a high-throughput sequencing method is used to successfully obtain a new Enterobacter cloacae 16SrDNA complete gene sequence SEQ ID NO.1 with the bacterial universal primer 27F / 1492R (Jiang and Wu et al., 2011), and then a large number of Primers, after analysis and screening, finally obtained a set of primers with excellent specificity and sensitivity, named Yg2. The sequence of the primer set is as follows:
[0055]
[0056]
[0057] 2. Establishment of LAMP amplification method
[0058] Using the isolated Enterobacter DNA as a template, using the positive and negative (water) of the kit (Guangzhou Double Helix Company) as a control, use the primer set Yg2 to refer to the following reaction system, and carry out LAMP amplification sequence at 63°C, such as SEQ ID Shown in NO.10. The amplified products were detected by ...
Embodiment 2
[0066] Specificity detection of embodiment 2 Yg2 primer set
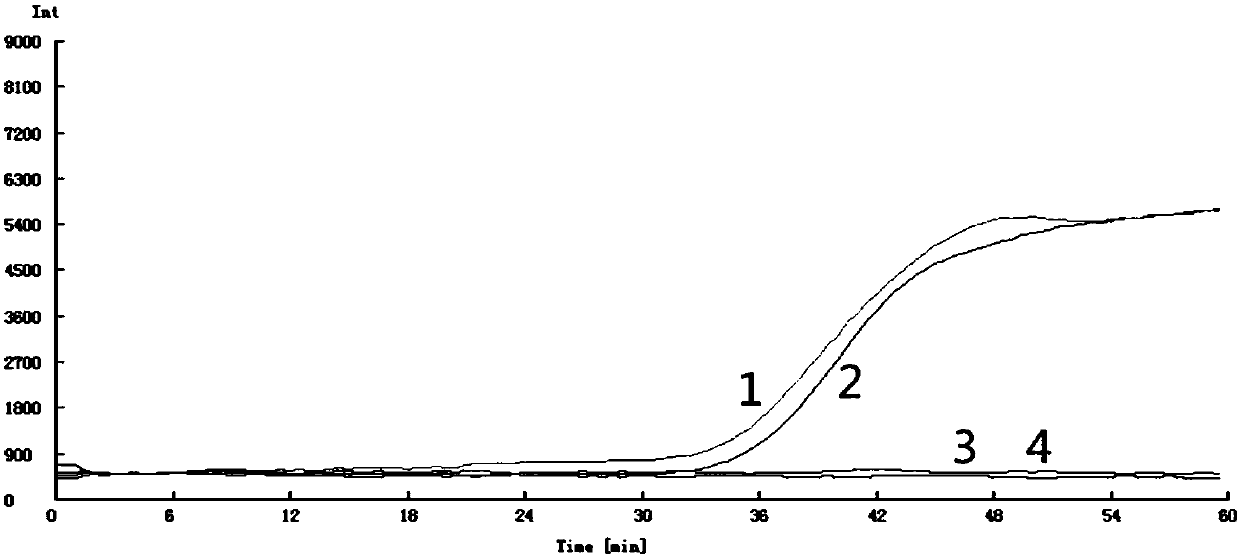
[0067] 1. Use a variety of bacteria such as Pseudomonas DNA, Bacillus DNA, and Klebsiella DNA isolated from the roots and stems of diseased plants, and Phyllactinia moricola, the pathogenic fungus of mulberry trees. ), mulberry bacterial wilt pathogen Laueria DNA, mulberry virus disease mosaic wilt pathogen DNA, mulberry leaf stain pathogen DNA and Beauveria bassiana DNA, Aspergillus nomius DNA, Penicillium DNA, Cladosporium DNA DNA and other fungi were used as a control group, and the yg2 primer set was used to perform fluorescence amplification, color reaction, and agarose gel electrophoresis detection by the method of Example 1.
[0068] 2. The amplification results of the primers are as attached Figure 4 , Figure 5 , Figure 6 shown. The results showed that only the DNA of the mulberry wilt pathogen (Enterobacter cloacae) showed an S-shaped curve at the target position, and the display results turned green...
Embodiment 3
[0069] Sensitivity detection of embodiment 3 Yg2 primer set
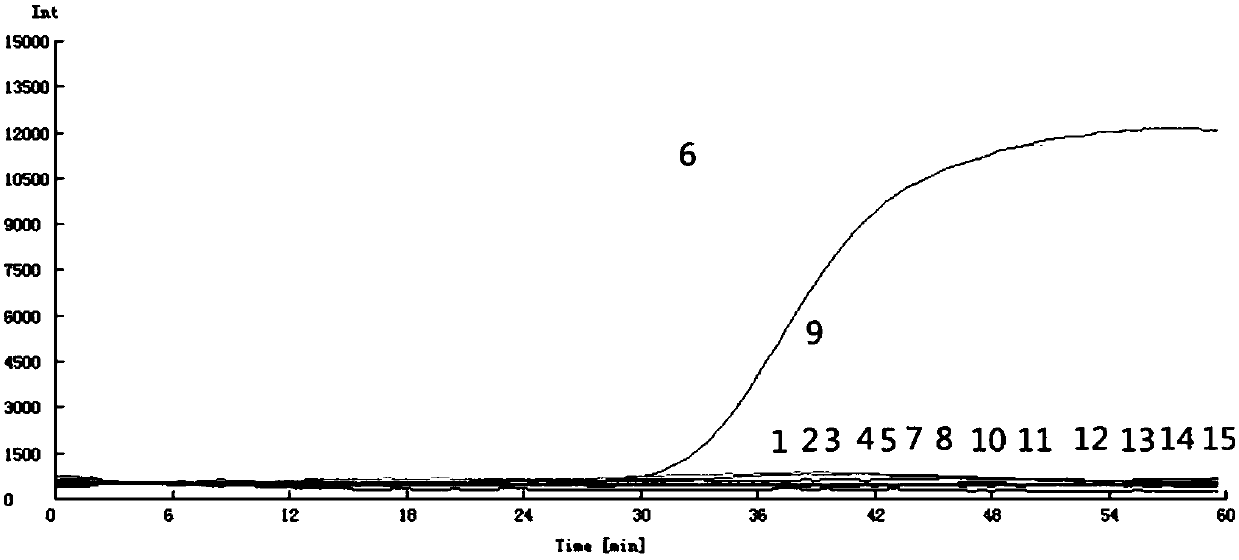
[0070] 1. Extract the DNA of Enterobacter cloacae, the original concentration is 50ng / μL.
[0071] Dilute the above DNA with 1×TE, dilute 10, 10 2 、10 3 、10 4 、10 5 、10 6 times. That is to say, the concentration gradient is 5.0, 5.0×10 -1 , 5.0×10 -2 , 5.0×10 -3 , 5.0×10 -4 , 5.0×10 -5 ng / μL.
[0072] 2. Using the above concentrations of DNA as a template and the Yg2 primer set, LAMP fluorescence amplification, color reaction, and agarose gel electrophoresis detection were carried out by the method of Example 1.
[0073] 3. The result is as follows Figure 7 , Figure 8 , Figure 9 As shown, the Yg2 primer set can detect the DNA concentration of pathogenic bacteria at 5.0×10 -4 , with good detection sensitivity.
[0074] 4. Culture to 1×10 8 CFU / mL bacterial liquid gradient dilution is 1×10 7 CFU / mL, 1×10 6 CFU / mL, 1×10 5 CFU / mL, 1×10 4 CFU / mL, 1×10 3 CFU / mL, 1×10 2 CFU / mL, 1×10 1 CFU / mL, 1×1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com