Rhizomucor miehei alpha-amylase, and encoding gene and application of rhizomucor miehei alpha-amylase
A technology of Rhizomucor miehei and amylase, applied in application, genetic engineering, plant gene improvement, etc., to achieve high-efficiency expression, delay aging, and good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Embodiment 1. Expression of Rhizomucor miehei α-amylase in recombinant Pichia pastoris
[0063] 1. Construction of recombinant bacteria
[0064] Design the upstream primer RmAmyAEcoRIF:
[0065] 5'-CCG GAATTC AAGCCATTGCCACTCGCTAAG-3' (the underline shows the EcoRI restriction site, and the sequence after the underline matches the 58th-78th positions in sequence 1 of the sequence listing);
[0066] Design the downstream primer RmAmyANotIR:
[0067] 5'-GAAT GCGGCCGC TTAAGCTCTCTGGAAAATAGCGGG-3' (the underline shows the Not I restriction site, and the sequence after the underline matches the 1375-1398th position in sequence 1 of the sequence listing);
[0068] Using the cDNA of Rhizomucor miehei α-amylase as a template, PCR amplifies the amino acid coding gene sequence of the protein. The PCR amplification conditions were: pre-denaturation at 95°C for 3 min, denaturation at 95°C for 30 s, annealing at 57°C for 30 s, extension at 72°C for 90 s, 34 cycles, and post-ext...
Embodiment 2
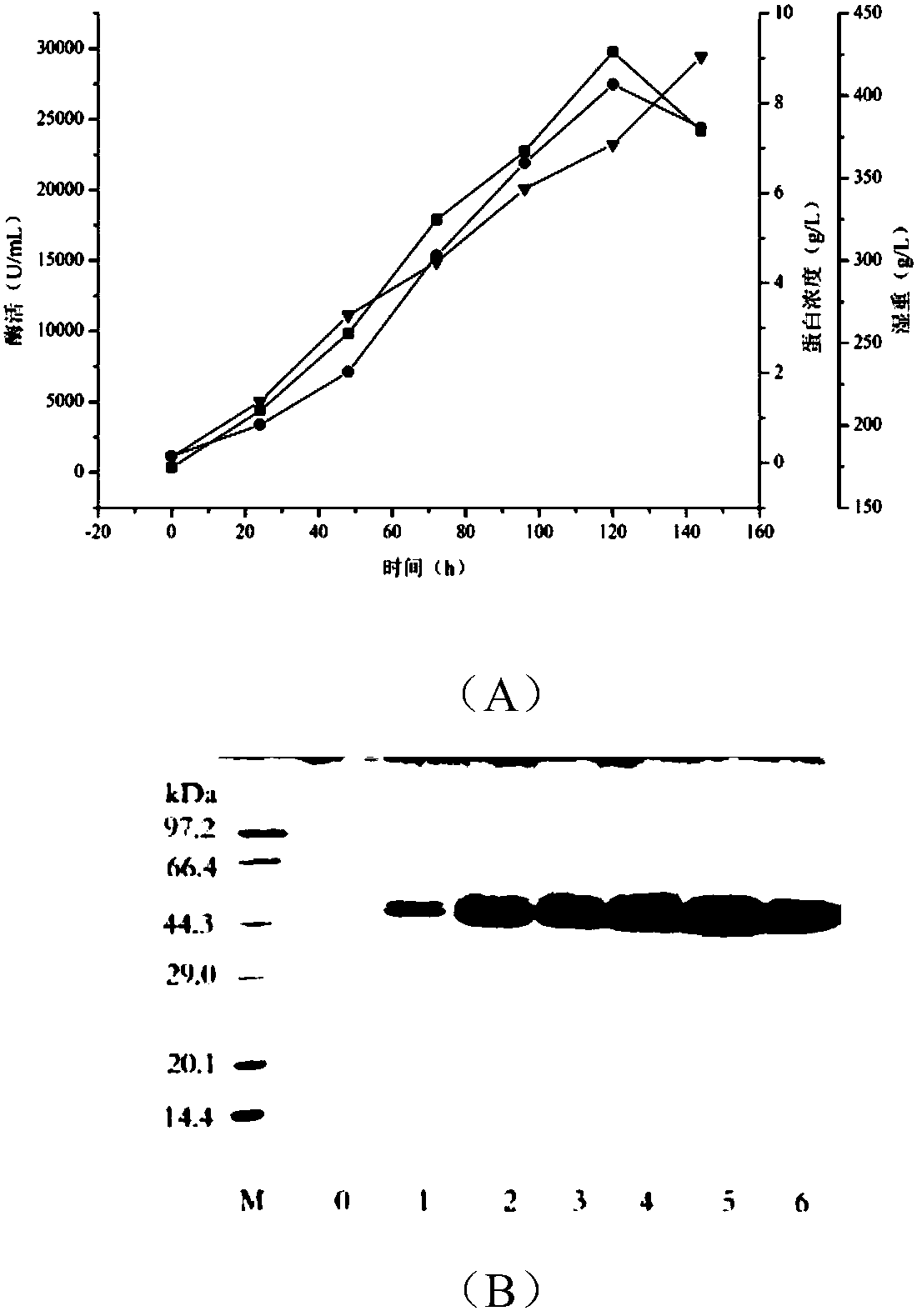
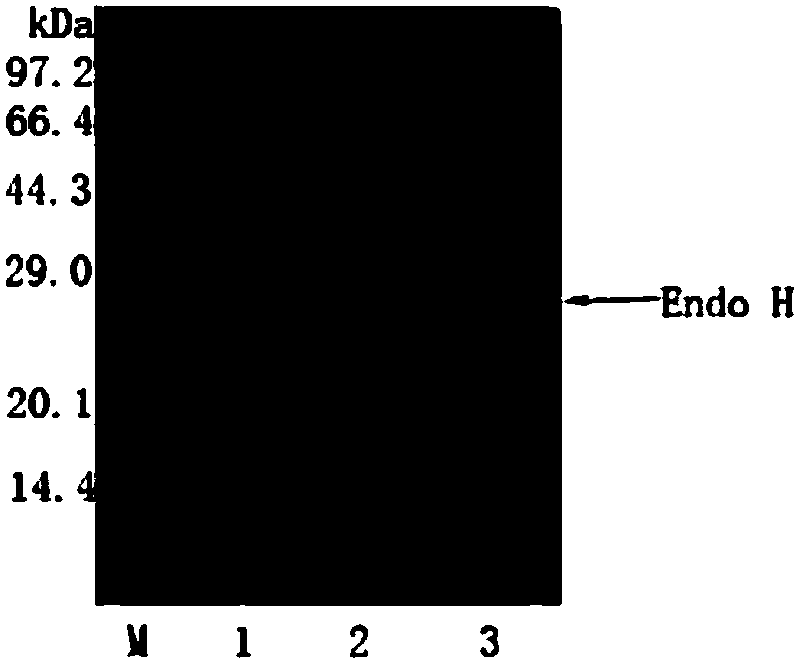
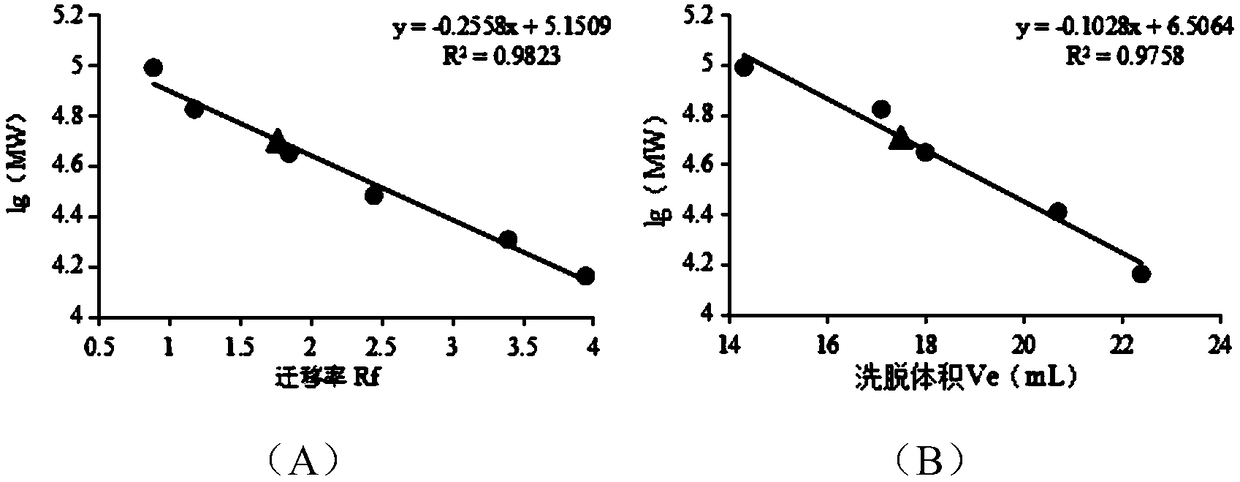
[0078] Example 2. Purification and enzymatic properties of α-amylase
[0079] 1. Purification of α-amylase
[0080] The fermentation broth was centrifuged at 4°C and 10000rpm for 10min, the supernatant was placed in 20mM PB (pH 6.0) buffer and dialyzed overnight at 4°C, and the dialyzed enzyme solution was centrifuged at 4°C and 10000rpm for 10min. The treated enzyme liquid was purified by DE52 weak anion column. Chromatography column was pre-equilibrated with 20mM PB (pH 6.0) buffer at a flow rate of 0.5mL / min; the enzyme solution was loaded at a flow rate of 0.5mL / min; the column was washed with 20mM PB (pH 6.0) buffer to OD 280 Less than 0.1, the flow rate is 1.0mL / min; wash the column with 20mM PB (pH 6.0) buffer containing 100mM NaCl to OD 280 Less than 0.1, the flow rate is 1.0mL / min; wash the column to OD with 20mM PB (pH 6.0) buffer containing 200mM NaCl 280 is less than 0.1, the flow rate is 1.0mL / min, collect the eluted solution, and use SDS-PAGE electrophoresis to ...
Embodiment 3
[0102] Example 3. Application of α-amylase in steamed bread
[0103] 1. The method of making steamed buns
[0104] Weigh 500g of flour and put it into the dough mixer, dissolve 4g of yeast in 235g of water, pour the water into the flour, stir at low speed to make the flour into a ball, and stir at high speed for 3 minutes. Divide the dough into 100g / piece, round the dough, put it in a proofing box at 38°C and 80% relative humidity for 45 minutes, and put it in a steamer for 15 minutes after proofing. The amount of RmAmyA added is 0.25-1.25 ppm (mg / Kg flour).
[0105] 2. Determination of aspect ratio and specific volume of steamed bread
[0106] Specific volume: After cooling the steamed buns at room temperature for 1 hour, weigh the mass of the steamed buns with a balance, and measure the volume of the steamed buns with the rapeseed row volume method. The specific volume of the steamed buns is the ratio of volume to mass.
[0107] Height-to-diameter ratio: Use a vernier cal...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com