3-mercaptopyruvate sulfurtransferase and application thereof
A technology of mercaptopyruvate sulfur transferase, applied in the field of molecular biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Embodiment 1. Arabidopsis AtMST Gene cloning.
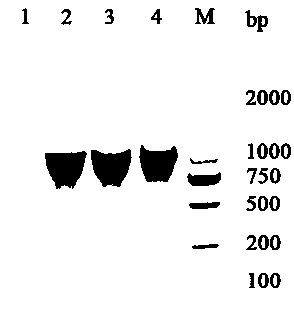
[0024] Take 4-week-old rosette leaves, extract total RNA (TRIzol Extraction Kit, TaKaRa Company), reverse transcribe into cDNA (Degenome Reverse Transcription Kit, abm Company), use the cDNA as a template, and use AtMST Specific primers for PCR, cloned AtMST Gene. Such as figure 1 It shows that after high-fidelity PCR using Arabidopsis cDNA as a template, a fragment with a size of 1140 bp is obtained as the target fragment; the negative control is ddH 2 O is the template, no bands were obtained; the fourth swimming lane is the recombinant pET28a- AtMST The fragments obtained from the bacterial liquid PCR detection of the construct indicated that the recombinant construct was constructed successfully.
Embodiment 2
[0025] Embodiment 2. T7:: AtMST Transformation of E. coli with recombinant plasmid, comparison experiment between E. coli induced by IPTG and E. coli without IPTG induction (method of preparing target protein and blank control experiment):
[0026] Preparation of experimental materials: extract total RNA from leaves of Arabidopsis thaliana, reverse transcribe into cDNA, clone AtMST gene (At1g79230), construct T7:: AtMST Recombinants were transformed into Escherichia coli BL21 (DE3) competent cells, and positive bacteria were obtained by kanamycin screening. A single colony was picked, identified by PCR, and successfully transformed Escherichia coli was obtained for experimentation.
[0027] The above two Escherichia coli were divided into two groups. One group was added with IPTG (final concentration: 0.05-0.5 mM), and the other group was added with an equal amount of double-distilled water, and cultured with shaking at 25-37°C for 12-20 h. Centrifuge at 10,000 rpm at 4°C ...
Embodiment 3
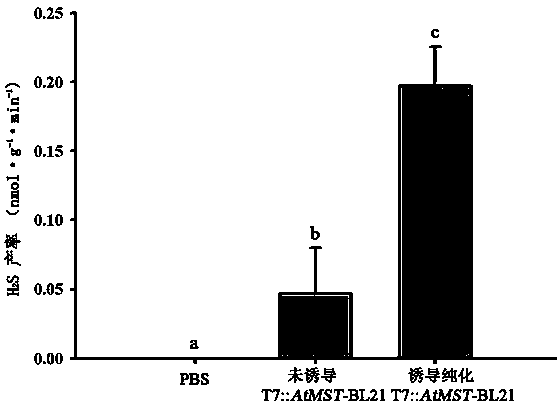
[0029] Example 3: H 2 Determination experiment of S yield.
[0030] Mix the reaction system according to the following formula: add 150 μmol·L -1 Sodium mercaptopyruvate, 100 μg protein, and the rest supplemented with PBS buffer solution with pH=7, the mixture was added to a 40 mL Erlenmeyer flask with a cover; the cap of a 1.5 mL centrifuge tube was cut off, and 500 μL of 1% zinc acetate solution; put the centrifuge tube into the Erlenmeyer flask and cover the Erlenmeyer flask cap. 37°C, shaking at 115 rpm, after fully reacting for 15 min, add 100 μL of 20 mM DPD and 30 mM FeCl respectively 3 The solution terminated the reaction, stood in the dark for 15 min, and measured the OD 670 . H was calculated from the standard curve 2 The yield of S.
[0031] image 3 The results showed that when induced by IPTG, AtMST The mercaptopyruvate sulfur transferase produced by gene expression can use sodium mercaptopyruvate as a substrate to catalyze H 2 S production, thereby incre...
PUM
| Property | Measurement | Unit |
|---|---|---|
| quality score | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com