InDel molecular marker closely linked with flowering time of photoperiod-sensitive Chinese cabbage crop and application thereof
A molecular marker, flowering time technology, applied in the field of genetic engineering and molecular biology, can solve the problems affecting plant phenotype, photoperiod sensitivity differences, etc., and achieve the goal of speeding up the breeding process, stabilizing variation, and improving screening efficiency and accuracy. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
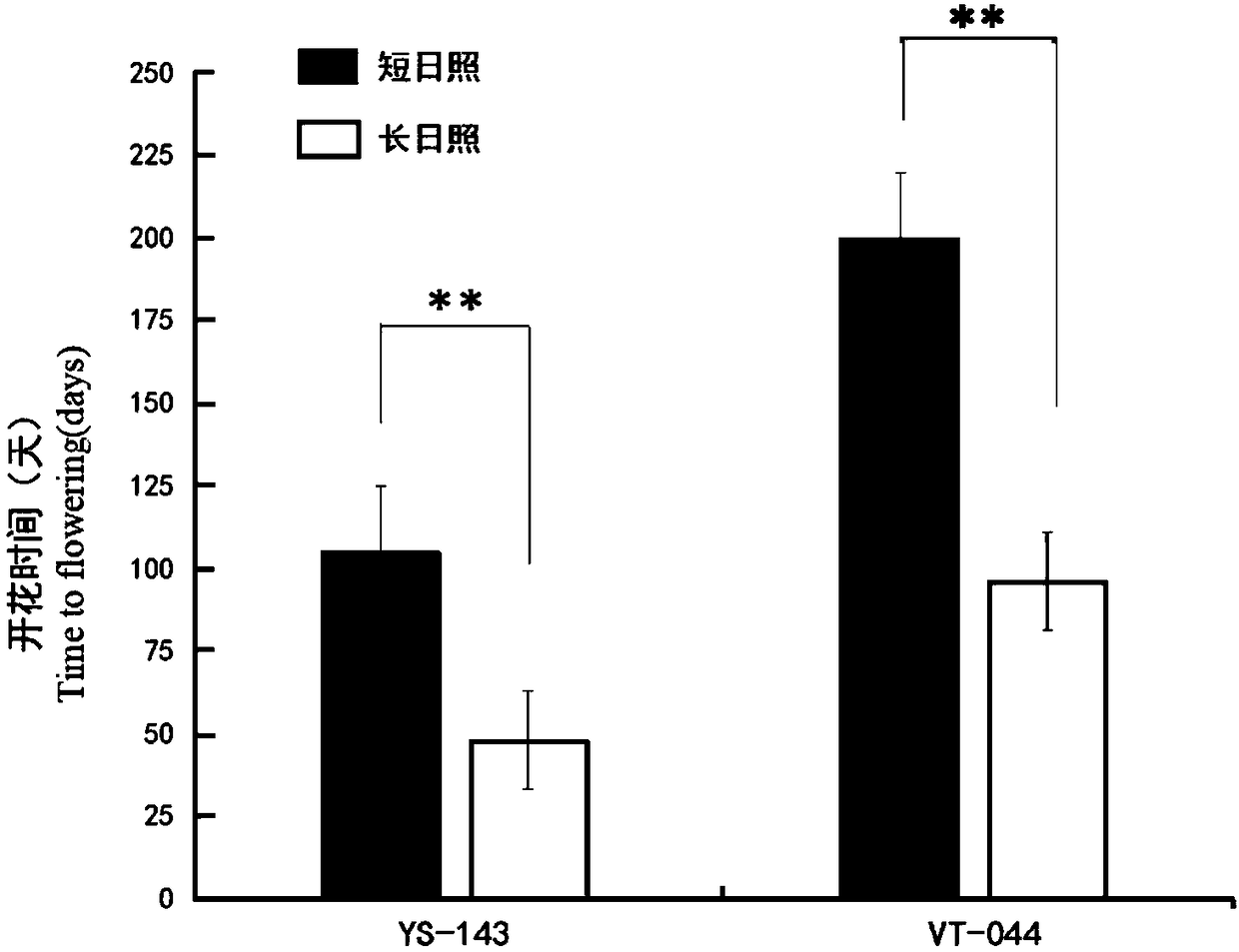
[0041] Embodiment 1 determines photoperiod sensitive material
[0042] On August 3, 2017, 20 seeds of the parent material were germinated in a petri dish covered with filter paper, and then sowed in a plug tray after 36 hours of dark cultivation at 25°C. Sunlight (light 8h / darkness 16h), temperature 22°C / 18°C, relative humidity 70%, light intensity 72μmol·m-2·s-1 grow in a light incubator, and plant pots (17cM) after one month. At the same time, the F 2 The 180 strains of the generation segregation population were also sown in the hole trays, and after 3 weeks of growth in plastic greenhouses, they were planted in plastic greenhouses in Jurong Agricultural Expo Park, Nanjing Agricultural University from August 24, 2017 to December 26, 2017. Individual plants are numbered sequentially and managed in the normal field. Observing the parent and F 2 The flowering time of the generation segregation population, the flowering time investigation standard is the time (number of days)...
Embodiment 2
[0045] Embodiment 2 determines candidate markers
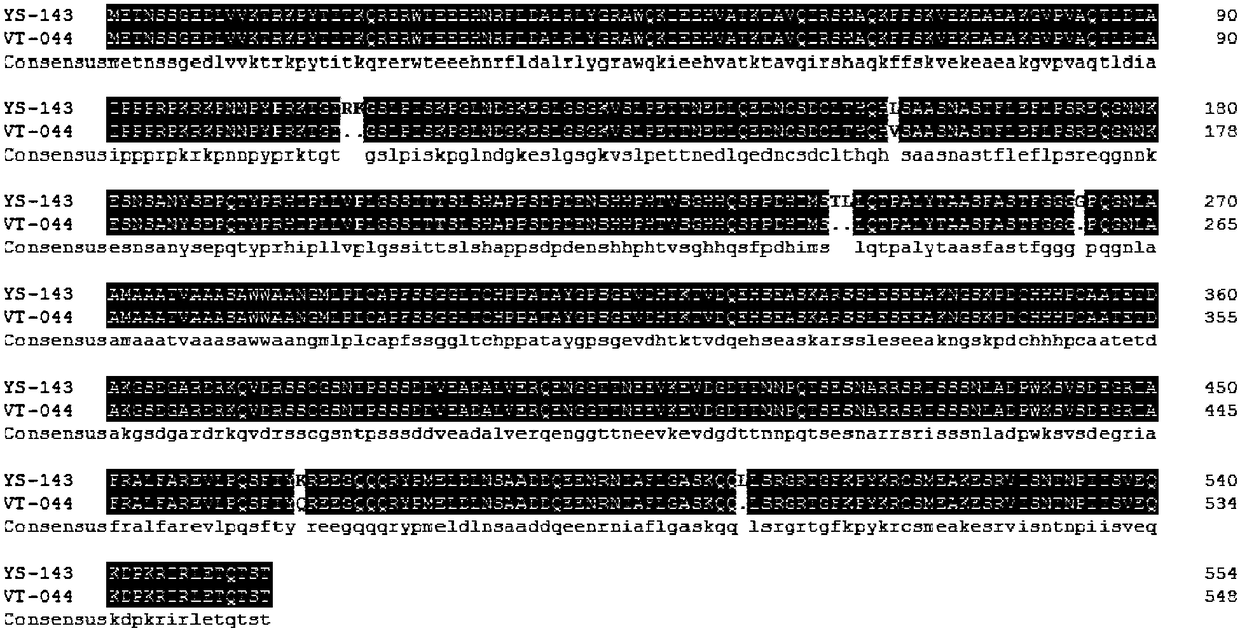
[0046] RNA Simple Total RNA Kit (TaKaRa) was used to extract the total RNA of the two parents. For specific methods, refer to the kit instructions. cDNA using PrimeScript TM II 1st Strand cDNA Synthesis Kit (TaKaRa) was synthesized by reverse transcription and used as a template for gene cloning.
[0047] CCA1 gene sequence refers to the published Chinese cabbage genome sequence (http: / / brassicadb.org / brad / :Bra004503; NCBI accession number: LOC103866427), using the online software Primer 3 (http: / / bioinfo.ut.ee / primer3- 0.4.0 / ) design the PCR primers for specific amplification of CCA1 gene,
[0048] CCA1-F: ATGGAGACTAATTCGTCTGGAG (SEQ ID NO. 7),
[0049] CCA1-R: TCATGTTGAAGTTTGTGTTTCC (SEQ ID NO. 8)
[0050] And synthesized by Nanjing Qingke Company.
[0051] The total volume of the PCR amplification system is 40 μL: 2 μL each of forward and reverse primers, 1 μL of template cDNA, 20 μL of high-fidelity enzyme MIX, and 15...
Embodiment 3
[0056] Embodiment 3 candidate mark and F 2 Correlation analysis and identification of flowering time in generation population
[0057] Extract F 2 The DNA of 24 leaves of the very early and very late flowering plants of the generation population, the specific method refers to the Axygen Plant Genomic DNA Extraction Kit.
[0058] Design specific primers pF1, pR1; pF2, pR2; pF3, pR3 (Table 1) for the 4 candidate markers (InDels) of the CCA1 coding region identified in the parents;
[0059] DNA from extremely early-flowering plants and DNA from extremely late-flowering plants were divided into 4 groups, a total of 8 groups, which were used as templates required for gene cloning of each differential site for PCR amplification. Here, 4 candidate markers and each group used The primers are:
[0060] Primer sequences used in the test in Table 1
[0061]
[0062] The total volume of the PCR amplification system in each group is 40 μL: 2 μL each of forward and reverse primers, 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com