Screening method for Fe<2+> and alpha-ketoglutaric acid-dependent hydroxylase and isoleucine hydroxylase
A technology of ketoglutarate dehydrogenase and ketoglutarate, which can be used in botany equipment and methods, biochemical equipment and methods, and other methods for inserting foreign genetic materials, and can solve the problem of high-activity hydroxylase screening methods Problems such as missing, hydroxyl compound development and application of industrial production constraints, to achieve the effect of improving catalytic efficiency, enzyme activity and thermal stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Construction of α-ketoglutarate dehydrogenase coding gene deletion and isocitrate lyase gene deletion strains
[0036] (1) Construction of homology arm overlapping gene fragments for knockout
[0037]According to the nucleotide sequence of sucA shown in Sequence Listing 6, its upstream and downstream homologous sequences were designed. The upstream and downstream gene fragments were obtained by DNA polymerase chain reaction. The upstream homology arm gene fragment and the downstream homology arm gene were used as templates, and sucA-up-1 and sucA-down-2 were used as upstream and downstream primers to perform polymerase chain reaction to obtain overlapping gene fragments of the upstream and downstream homology arms. After purification by 1.0% agarose electrophoresis, it was purified with a gel cutting recovery kit. in,
[0038] sucA upstream homology arm sequence primer:
[0039] sucA-up-1: 5'TCTGGTGGAATCCCGATAAGTT 3'
[0040] sucA-up-2: 5'TTACGACGCTGTTCCTGTTTCTATCC...
Embodiment 2
[0058] Recombinant expression vector containing mutant and construction of strain containing the vector
[0059] Using the isoleucine hydroxylase coding gene ido as a template, the sequence is shown in Sequence Listing 4, and pWSK29-1 and pWSK29-2 were used as primers to perform error-prone PCR, and the product was purified after 1.0% agarose electrophoresis Afterwards, the ido gene and its mutant fragments are obtained by purifying with a gel-cutting recovery kit. Primer sequences are as follows
[0060] pWSK29-1:5'
[0061] AGGGAACAAAAGCTGGAGCTCGAAGGAGATATACAAATGAAAATGAGCGGTTTTTAGCAT 3'
[0062] pWSK29-2:5'
[0063] GGGCCCCCCCTCGAGGTCGACTTATTTGGTTTCTTTATAGCTAAAGGTCA 3'
[0064] The obtained above ido gene fragments were recombined and ligated with the pWSK29 vector after digestion with Sac I and Sal I restriction endonucleases in a ligation system containing Exnase II, and the ligated reaction product was transformed into host cell E.coli From W3110△sucA△aceA competent ...
Embodiment 3
[0066] hyperactive ido mutant ido M screening
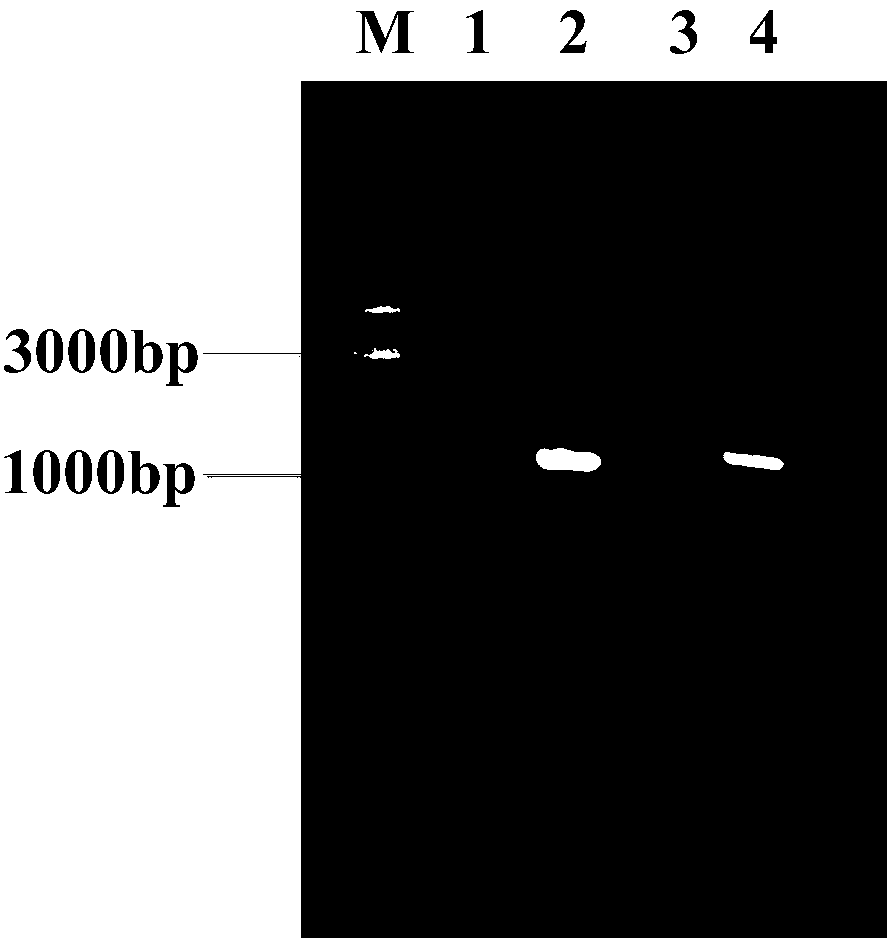
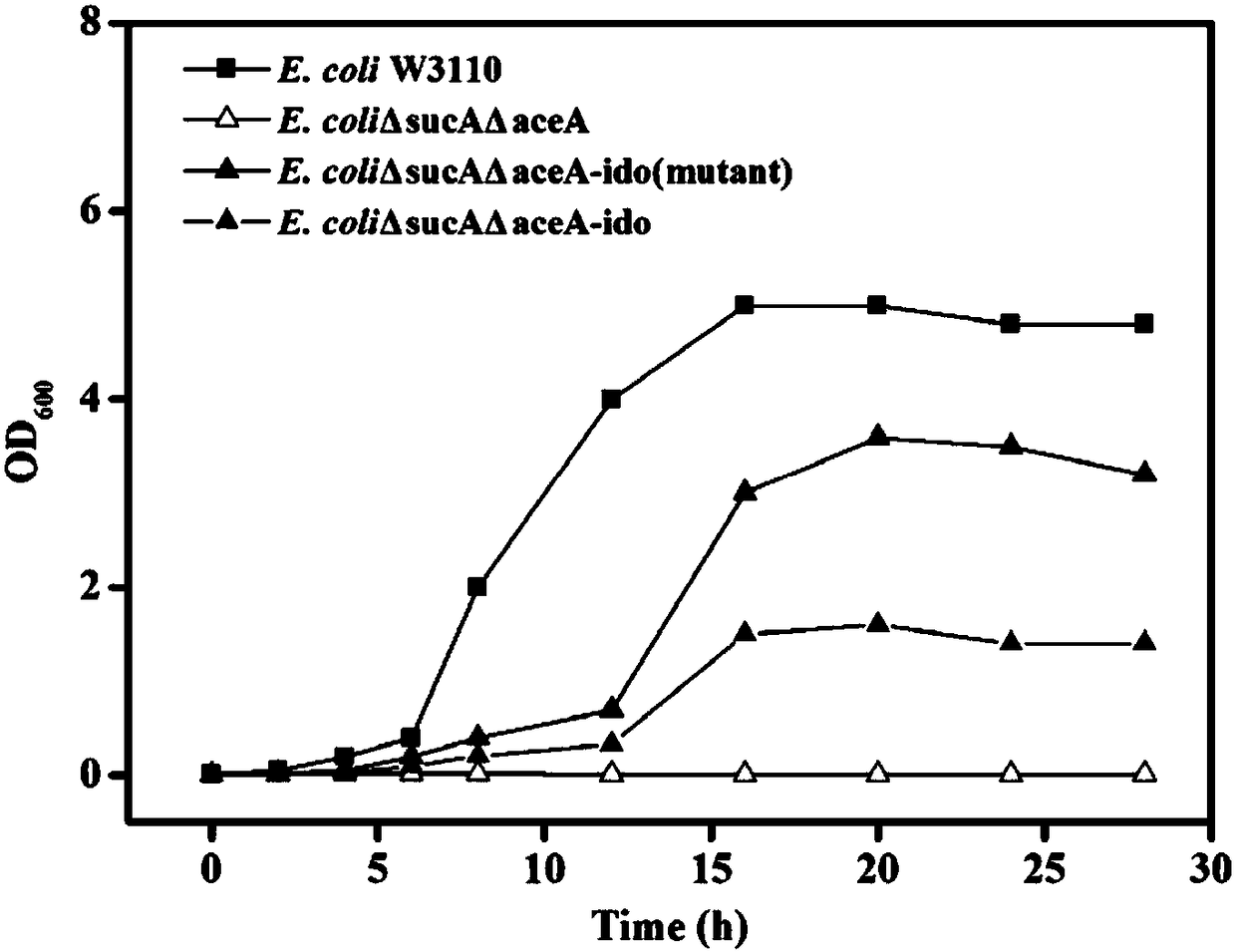
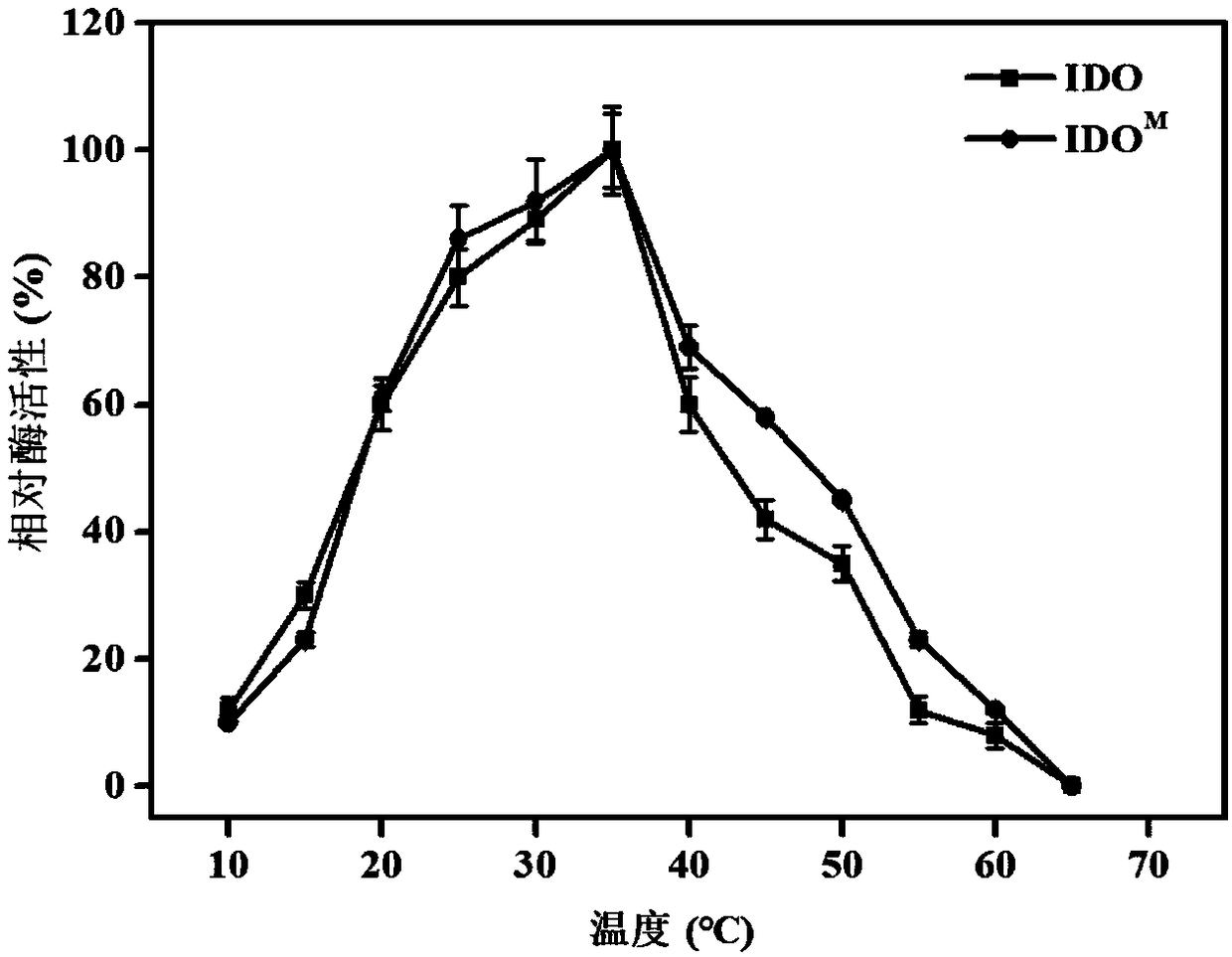
[0067] The bacterial strain containing the mutant recombinant expression vector obtained in Example 2 was applied to solid selection medium (glucose 10g / L, MgSO 4 0.24g / L, Na 2 PO 4 ·7H 2 O 13g / L, KH 2 PO 4 2.5g / L, NH 4 SO 4 5g / L, Isoleucine 2g / L, FeSO 4 2g / L, Vc 0.2g / L, agar 20g / L, tap water 1000mL, pH 6.5-7.5) plate, cultured upside down at 37°C for 12 hours, pick 480 larger single colonies on the plate and transfer to 5 blocks containing ampicillin (100μg / mL) liquid screening medium (glucose 10g / L, MgSO 4 0.24g / L, Na 2 PO 4 ·7H 2 O 13g / L, KH 2 PO 4 2.5g / L, NH 4 SO 4 5g / L, Isoleucine 2g / L, FeSO 4 2g / L, Vc 0.2g / L, tap water 1000mL, pH 6.5-7.5) in a 96-well plate, cultured with shaking at 37°C for 24h. Determination of OD by microplate reader 600 . Select the bacterial strain with the highest OD600 and determine the growth curve, such as figure 2 shown. Extract the plasmid and its ido mutant ido M P...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com