Method of researching gene function of Bradyrhizobium japonicum USDA110
A brachyrhizobia and gene technology, applied in the field of microorganisms, can solve the problems of researchers' difficulties, restrict the research on the function of brachyrhizobia genes, lack of suitable research methods, etc. The effect of speeding up the progress of experimental research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1 Preparation of Pseudomonas aeruginosa PAO1 Electric Shock Competent Cells
[0064] The preparation of Pseudomonas aeruginosa PAO1 electric shock competent cells comprises the following steps:
[0065] (1) Pseudomonas aeruginosa PAO1 was shaken overnight for 8 hours, 6 mL was taken as a competent state, and divided into three 2 mL sterilized centrifuge tubes, room temperature was 10,000 rpm for 2 minutes, and the supernatant was removed;
[0066] (2) Resuspend with filter-sterilized 10% sucrose, wash once, room temperature 10000rpm, 2min, remove supernatant;
[0067] (3) Repeat step 2 twice;
[0068] (4) Concentrate into 100 μl with 10% glycerol and use for electroporation of the constructed plasmid.
Embodiment 2
[0069] Example 2 Construction of bradyrhizobium USDA110 bll5123 gene overexpression strain and colony phenotype observation
[0070] 1. Extraction of DNA
[0071] Genome kit (EE161-01) from Beijing Quanshijin Biotechnology Co., Ltd. was used to extract the whole genome of Bradyrhizobium USDA110.
[0072] 2. Amplification of the bll5123 gene
[0073] Utilize NEB high-fidelity Q5 polymerase, carry out following PCR amplification, obtain containing bradyrhizobium USDA110 bll5123 (Gene ID: 1051644, namely NC_004463.1:c5683715-5682342 Bradyrhizobium japonicum USDA110 chromosome, complete genome) gene product, the amplified product was connected to a cloning vector and transformed into Escherichia coli for sequencing after culture, the positive strains were retained, and the plasmid was extracted.
[0074] According to NCBI records, the nucleotide sequence of USDA110 bll5123 is shown in SEQ ID NO: 1, which is:
[0075] GTGTCCGCGCGTATCCTGGTTGTCGATGACGTTCCTGCCAACGTCAAACTCCTCGAGGCC...
Embodiment 3
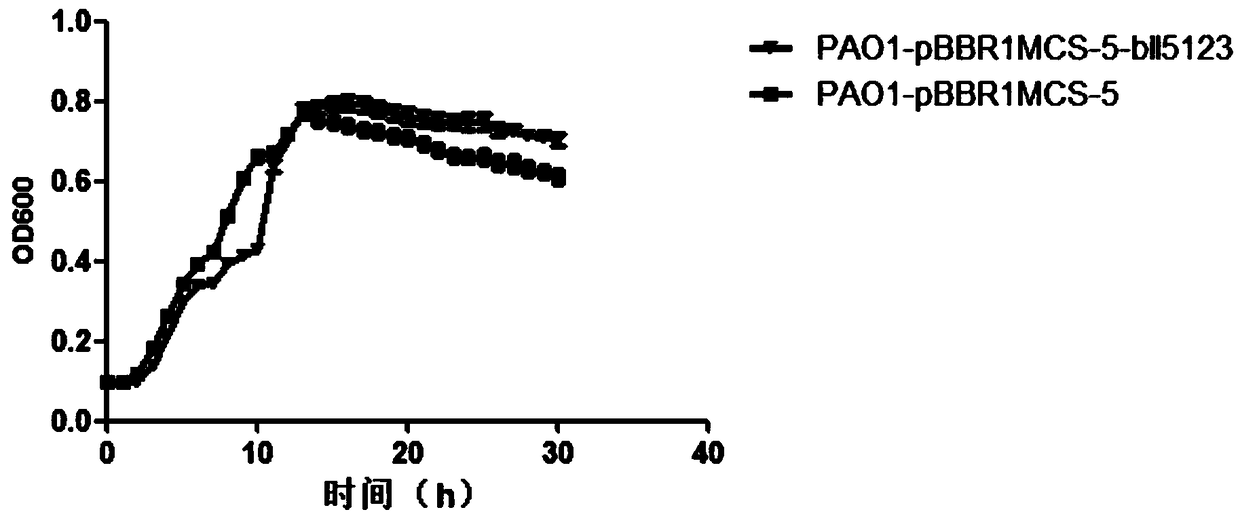
[0092] Example 3 Determination of Growth Curve
[0093] 1. Shake the transformants PAO1-pBBR1MCS-5 and PAO1-pBBR1MCS-5-bll5123 constructed in Example 2 overnight for 8 hours (LB gentamicin 50 μg / mL), and then add LB gentamicin at a ratio of 1:100 Add 200 μl of damycin 50 μg / mL liquid to the plate for measuring the growth curve, and detect it with the instrument for measuring the growth curve. There are 8 replicates in each group, and the experimental results are repeated three times.
[0094] 2. Results
[0095] As determined by the growth curve, figure 2 It was shown that the strain PAO1-pBBR1MCS-5-bll5123 overexpressing Bradyrhizobium USDA110 bll5123 and the control strain PAO1-pBBR1MCS-5 had no effect on the growth rate, and the subsequent experiments excluded the effect of growth rate.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com