Recombinant bacillus amyloliquefaciens as well as construction method and application thereof
A technology of amylolytic spores and construction methods, applied in the field of recombinant Bacillus amyloliquefaciens and its construction, can solve the problems of low price and high cost of polyglutamic acid degrading enzymes, achieve great application advantages and save production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
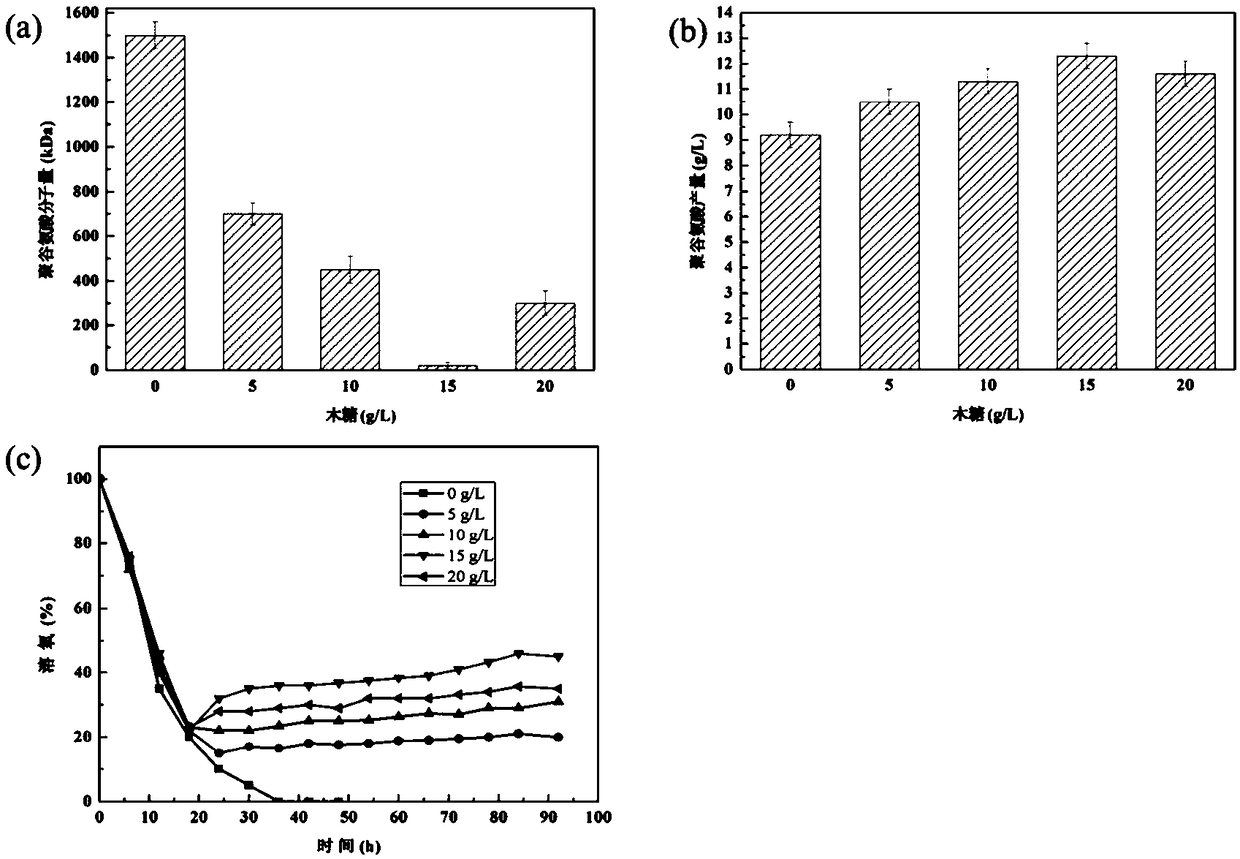
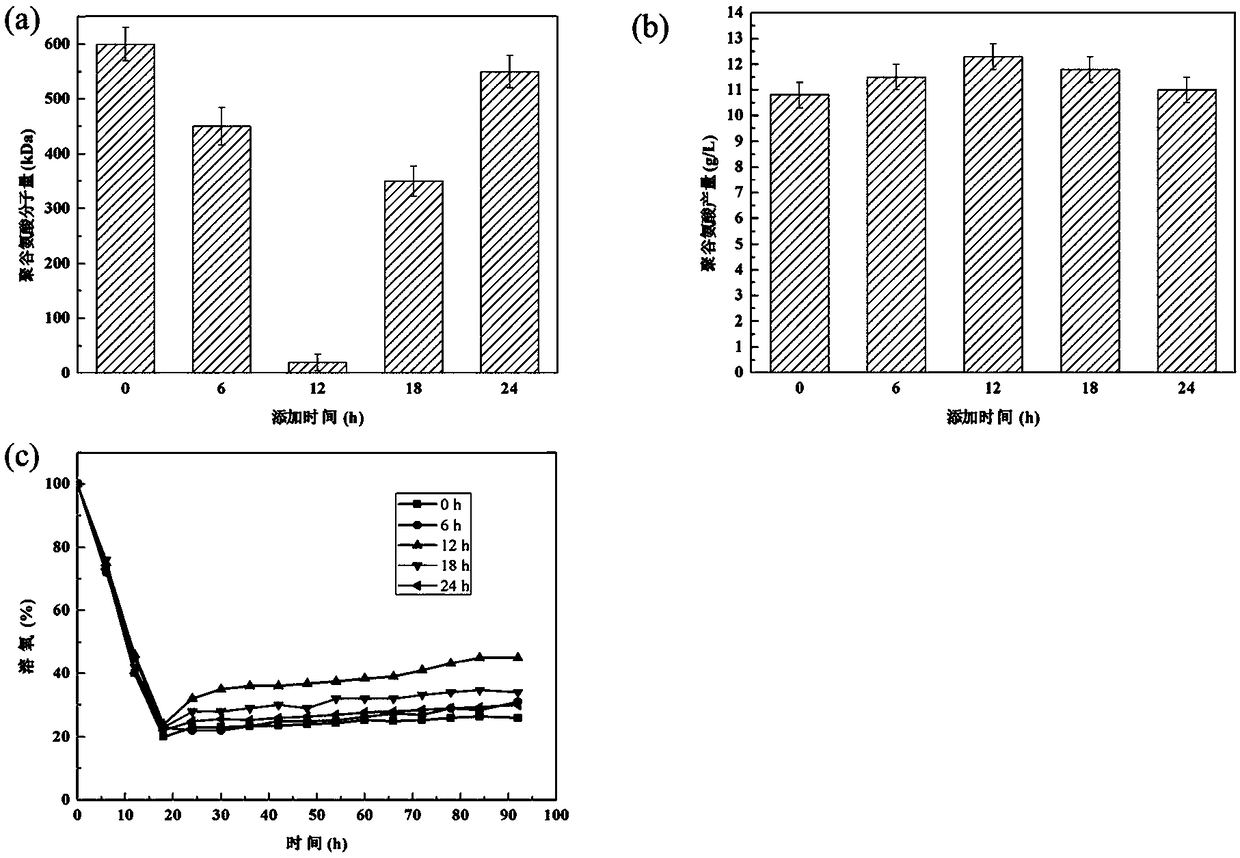
Image
Examples
Embodiment 1
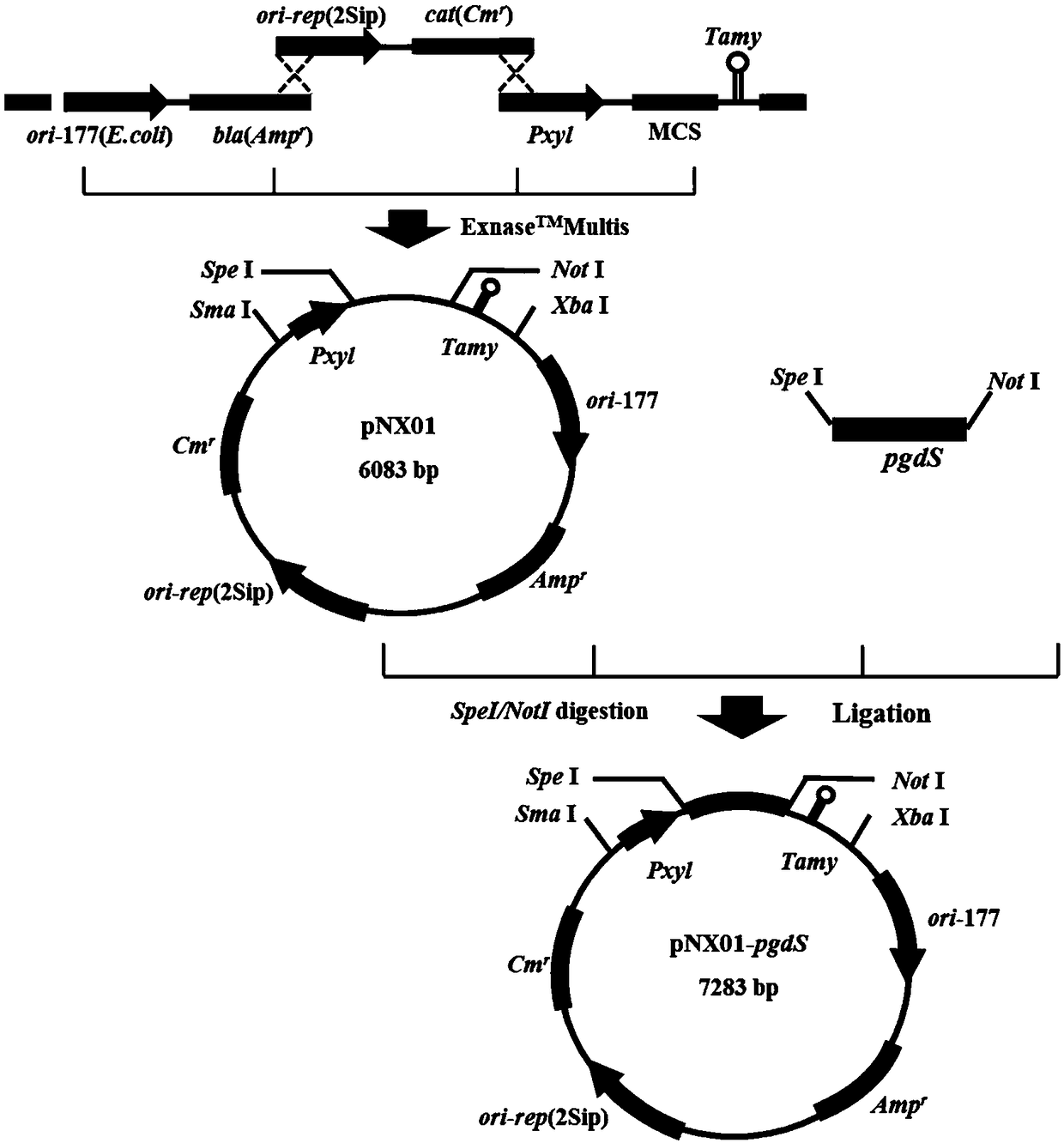
[0059] Construction of embodiment 1 plasmid pNX01
[0060] (1) Construction of vector skeleton Frag1
[0061] Using the shuttle plasmid pHY300PLK (TaKaRa Code No.3060) as a template, the origin of replication ori-177 of Escherichia coli and the ampicillin selection marker (such as SEQ ID NO: 1) were obtained, and the primer ori-F: 5'--3'( Such as SEQ ID NO: 10) and Amp-R: 5'--3' (such as SEQ ID NO: 11) for PCR amplification, the reaction conditions are shown in Table 1, using the DNA gel recovery kit (AP -MN-P-250G) recover the fragment to obtain the vector backbone Frag1.
[0062] Table 1 PCR reaction system and reaction conditions of Frag1
[0063]
[0064] (2) Construction of vector skeleton Frag2
[0065] According to the replication protein sequence (as shown in SEQ ID NO: 2) of the endogenous plasmid of B. amyloliquefaciens NX-2S (Bacillus amyloliquefaciens CCTCC NO: M 2016346) that has been determined, design primer rep-F respectively: (as shown in SEQ ID NO :12)...
Embodiment 2
[0091] Construction of embodiment 2 recombinant plasmid pNX01-pgdS
[0092] The polyglutamic acid degrading enzyme gene PgdS is derived from Bacillus subtilis B. subtilis NX-2, and the primers PgdS- F: (such as SEQ ID NO: 20) and PgdS-R: (such as SEQ ID NO: 21), use the bacterial genomic DNA extraction kit (TIANamp Bacteria DNA Kit) provided by Tiangen Biochemical Technology Co., Ltd. to extract the genome to extract The genome of pgdS was used as a template, and a standard PCR amplification system and program was used (see Table 9 for PCR reaction conditions) to obtain a pgdS gene fragment, the nucleotide sequence of which was shown in SEQID NO:9.
[0093] Table 9 PCR reaction system and reaction conditions of polyglutamic acid degrading enzyme gene PgdS
[0094]
[0095] The plasmid pNX01 was double-digested with Spe I and Not I, and the enzyme digestion system and reaction conditions are shown in Table 10;
[0096] Table 10 Plasmid pNX01 digestion reaction system and r...
Embodiment 3
[0101] Example 3 Pretreatment of recombinant plasmid pNX01-pgdS--demethylation
[0102] The recombinant plasmid pNX01-pgdS and E.coli GM2163 (Shanghai Haoran Biotechnology Co., Ltd. No. M0099) (competent mixing, and the same heat shock transformation method as in step (4) of Example 1 were used for the sequencing comparison. The plasmid was transformed into GM2163, and the transformants were picked for enzyme digestion verification again. The verified correct plasmid could be directly used as the plasmid to be electroporated because it did not have a BamH I site and avoided the recognition and cutting of the Bacillus amylobacter restriction repair system.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com