Recombinant bacteria for producing beta-alanine as well as construction method and application of recombinant bacteria
A construction method and technology of recombinant bacteria, applied in the biological field, can solve the problems of environmental pollution, unsuitable separation and purification, harsh reaction conditions of beta-alanine, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
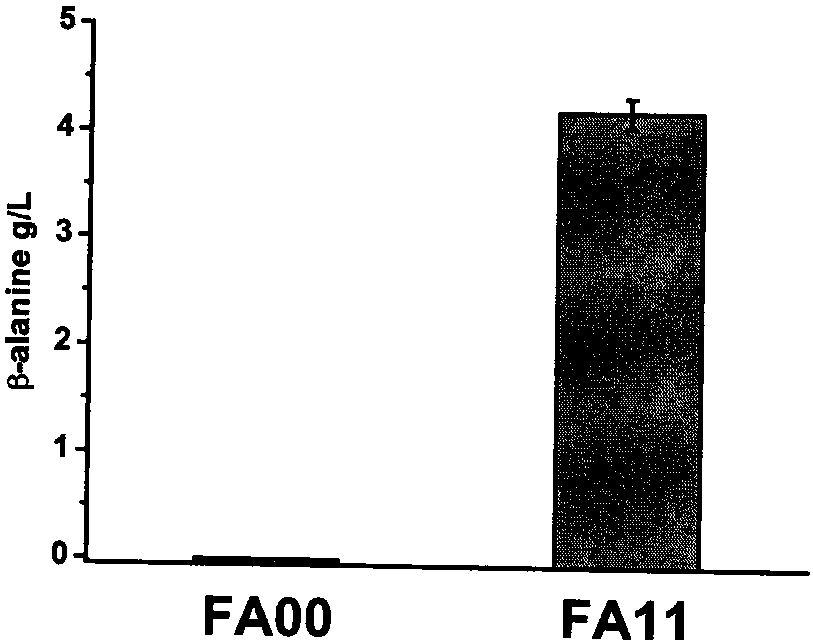
[0180] Embodiment 1, the construction of recombinant escherichia coli engineering strain FM07
[0181] In this example, a basic strain FM07, which can be used to prepare strains producing β-alanine and 3-hydroxypropionic acid, was prepared. The preparation method of the strain is as follows, and the primers used are shown in Table 1.
[0182] (1) Knockout of fatty acid degradation transcription factor fadR.
[0183] Starting from Escherichia coli BW25113, the fadR gene of Escherichia coli BW25113 was knocked out to obtain the mutant FM01 of Escherichia coli BW25113, the specific steps are as follows:
[0184] (1-a) Prepare a P1 phage containing an Escherichia coli gene fragment having a fadR knockout property.
[0185] The Escherichia coli gene fragment containing the fadR knockout trait comes from Escherichia coli strain JW1176, which is a W3110 series strain containing the fadR knockout trait. JW1176 is a product of the National Institute of Genetics (NIG, Japan) in Japan, ...
Embodiment 2
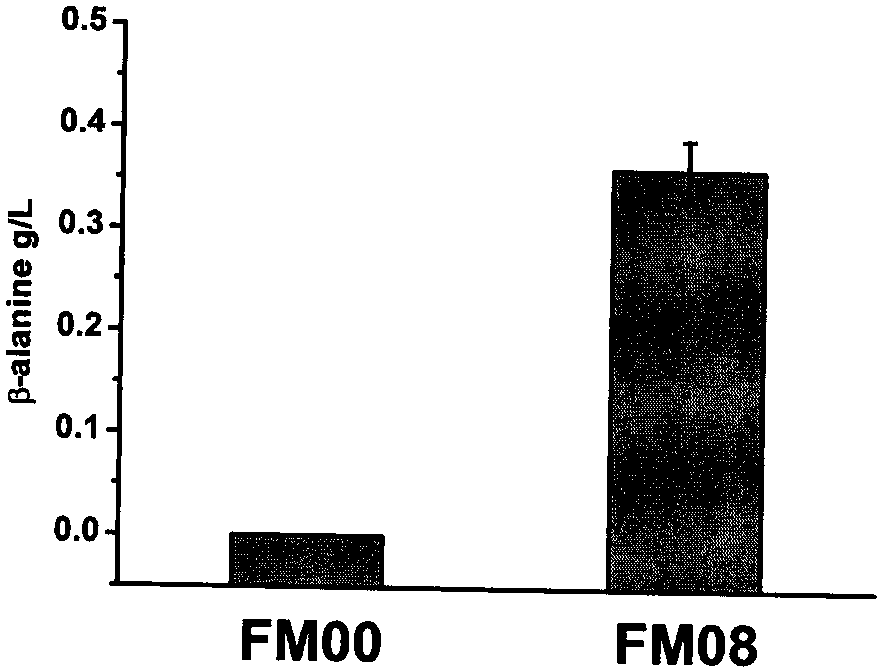
[0257] Embodiment 2, the preparation of the bacterial strain FM08 that is used to produce β-alanine and the production of β-alanine
[0258] One, for the preparation of the bacterial strain FM08 of producing β-alanine (β-alanine)
[0259] The preparation method of FM08 is as follows, and the primers used are shown in Table 2.
[0260] (1) Construction of a plasmid expressing a truncated gene mcrC of Chloroflexus aurantiacus malonyl-CoA reductase.
[0261] (1-a) PCR amplification of mcrC gene.
[0262] The nucleotide sequence of the transformed Chloroflexus aurantiacus malonyl-CoA reductase truncated gene mcrC is as SEQ ID No.22, and SEQ ID No.23 in the coding sequence list protein shown. Whole gene synthesis of the mcrC gene shown in SEQ ID No.22, and then using the Gibson assembly method (Gibson DG, YoungL, et al.Enzymatic assembly of DNA molecules up to several hundred kilobases.Nat.methods.2009; 6(5):343 -345) The mcrC gene shown in SEQ ID No.22 was connected to the pUC...
Embodiment 3
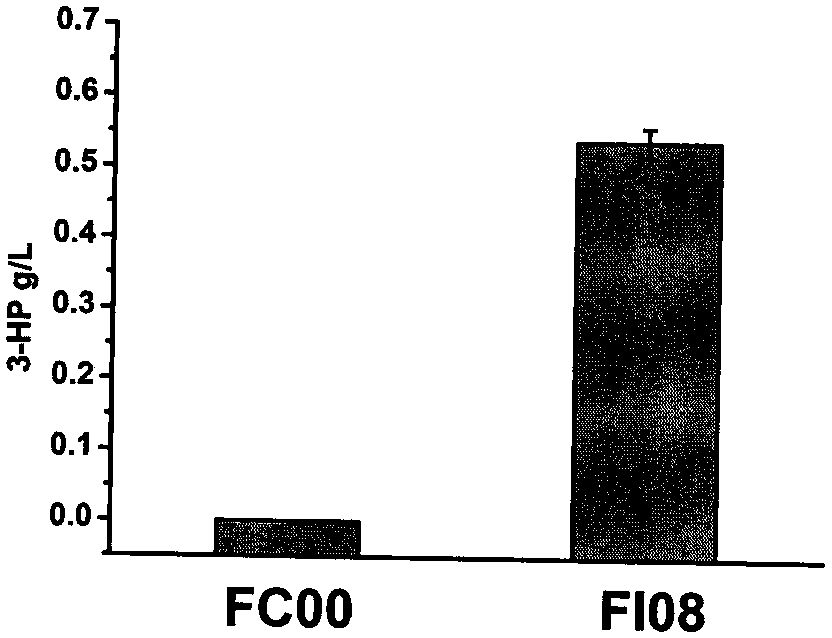
[0309] Embodiment 3, the preparation of bacterial strain FI08 for producing 3-hydroxypropionic acid and the production of 3-hydroxypropionic acid
[0310] 1. Preparation of bacterial strain FI08 for the production of 3-hydroxypropionic acid
[0311] The preparation method of FI08 is as follows, and the primers used are shown in Table 3.
[0312] (1) Construction of a plasmid expressing the acetyl-CoA carboxylase acc gene cluster of Corynebacterium_glutamicum.
[0313] (1-a) Extraction of genomic DNA of Corynebacterium glutamicum and PCR amplification of the acc gene cluster.
[0314] Genomic DNA of Corynebacterium glutamicum was extracted using a bacterial genome extraction kit (Tiangen Biochemical Technology Co., Ltd., product catalog DP302). Using the total genomic DNA extracted from Corynebacterium glutamicum as a template and accBC-F and accL-R as primers, the gene fragment accBC was amplified by high-fidelity TransStart FastPfu DNA polymerase PCR, and the target fragmen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com