Kit for detecting pathogenic vibrios
A pathogenic vibrio and kit technology, applied in the field of kits for detection of pathogenic vibrio, can solve the problems of high detection limit, long detection time, prone to false positives, etc., achieve high specificity and sensitivity, The effect of improving detection sensitivity and avoiding non-specific amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
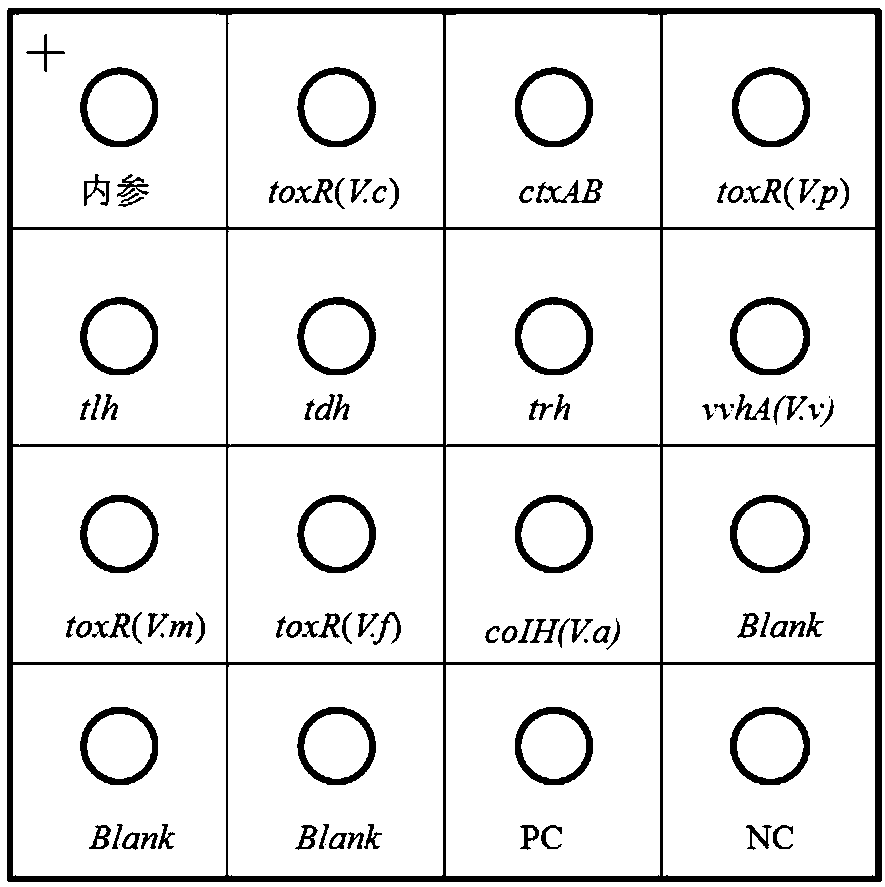
[0147] This embodiment describes a kit for detecting pathogenic Vibrio, which includes a nucleic acid combination and a membrane chip.
[0148] Wherein, the nucleic acid combination includes the following primer pairs, and the base sequences (5'-3') of the upstream primer and the downstream primer of each primer pair are as follows:
[0149] First primer pair for Vibrio cholerae toxR gene (toxR(V.c)):
[0150] Upstream primer: ATTGACGGCTACGCCATCGACA (SEQ ID NO.1), downstream primer: ACCGCAGCCCAGCCAATGTTG (SEQ ID NO.2);
[0151] Second primer pair for detection of Vibrio cholerae ctxAB gene:
[0152] Upstream primer: CATCTGGATGAGGACTGTATGC (SEQ ID NO.3), downstream primer: GCCAAGAGGACAGAGTGAGTA (SEQ ID NO.4);
[0153] The third primer pair for detecting Vibrio parahaemolyticus toxR gene (toxR(V.p)):
[0154] Upstream primer: GAAGGCAGCCAGATGTTGATT (SEQ ID NO.5), downstream primer: TCTCAGTTCCGTCAGATTGGT (SEQ ID NO.6);
[0155] The fourth primer pair used to detect the tlh gen...
Embodiment 2
[0206] The method for detecting pathogenic Vibrio in the sample to be tested by using the above kit is as follows:
[0207] 2.1 Use 3% sodium chloride alkaline peptone water to carry out mixed bacterial enrichment culture on the sample, and take the cultured mixed bacterial solution for DNA extraction; this step is carried out as required.
[0208] 2.1 Extract the genomic DNA of the sample to be tested according to the CTAB method, and use an ultra-micro spectrophotometer to measure the mass concentration of the DNA solution for later use.
[0209] 2.2 Multiplex PCR system and conditions
[0210] The reaction system of PCR amplification:
[0211] 10×PCR Buffer (with Mg 2+ , containing UNG enzyme): 5 μL;
[0212] dNTP (2.5mM each): 5μL;
[0213] Taq DNA polymerase (5U / μL): 0.5 μL;
[0214] Biotin-labeled downstream primers (20 μM): 1.2 μL; upstream primers (20 μM): 1 μL; instructions: 11 primer pairs (provided in Example 1) were added, each primer pair had 1.2 μL of downst...
Embodiment 3
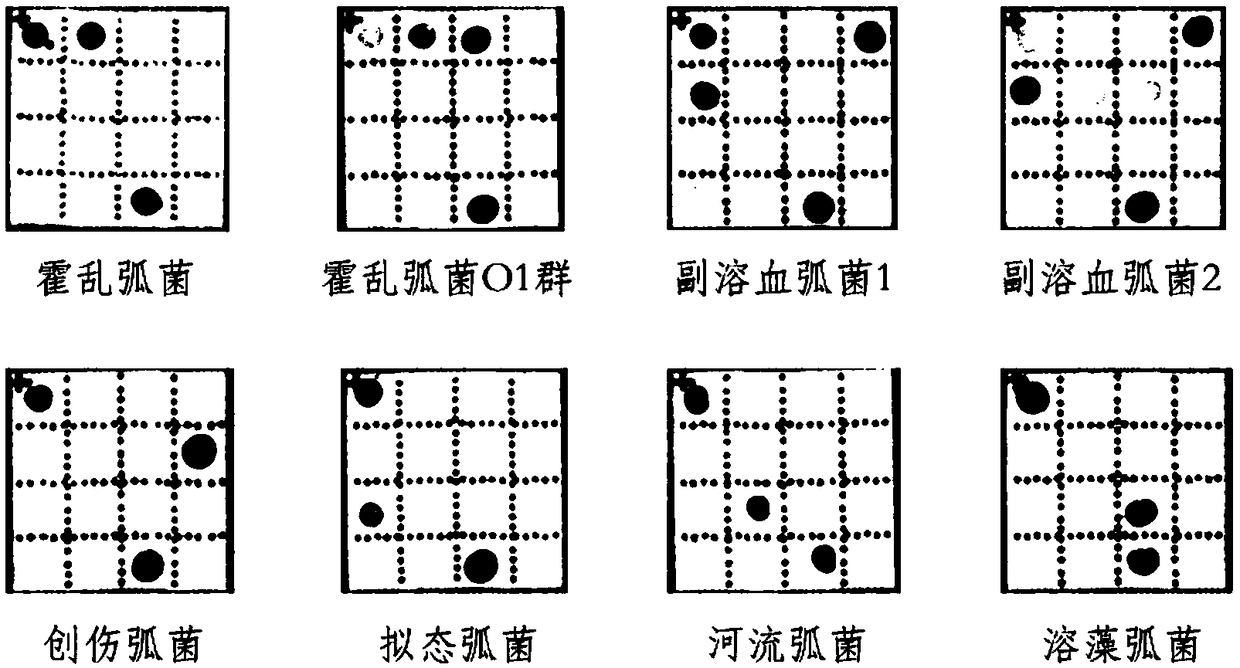
[0249] According to the instructions of Tiangen Biochemical Technology (Beijing) Co., Ltd. Bacterial Genomic DNA Extraction Kit (Cat. No.: DP302), extract different pathogenic Vibrio standard strains (Vibrio cholerae (without virulence factor ctxAB), Vibrio cholerae O1 group) (with virulence factors ctxAB), Vibrio parahaemolyticus 1 (without virulence factors tdh or trh), Vibrio parahaemolyticus 2 (with virulence factors tdh and trh), Vibrio vulnificus, Vibrio mimicus, Vibrio riverines and Vibrio alginolyticus) genomic DNA. The concentration and quality of the extracted DNA were measured with an ultra-micro spectrophotometer (Thermo NanoDrop 2000) for later use.
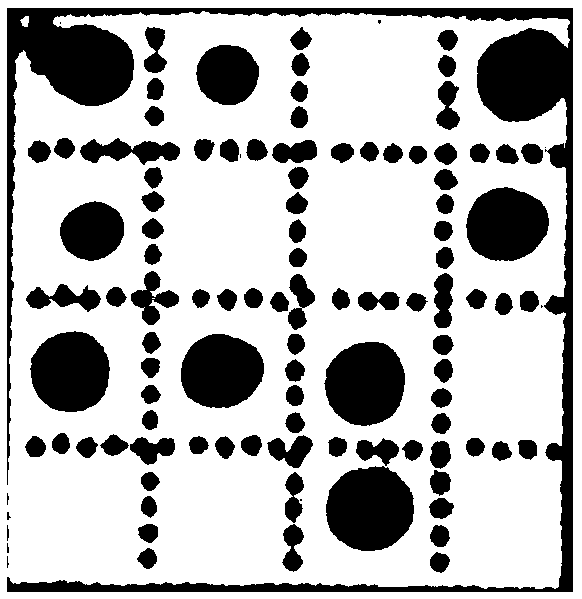
[0250] With the kit of Example 1, detect according to the method of Example 2, according to the color development of the hybridization point with reference to figure 1 Interpret the results, the results are as follows figure 2 shown.
[0251] figure 2 Vibrio cholerae (non-V. cholerae O1 or V. cholerae O139 grou...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com