Coding genes and applications of alginate lyases
A technology of alginate lyase and coding gene, which is applied in the direction of lyase, application, genetic engineering, etc., can solve the problems that are not suitable for large-scale industrial application and poor thermal stability, and achieve the purpose of improving industrial application value and thermal stability Good performance and high catalytic efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1, the extraction of the genomic DNA of the marine thermophile Defluviitalea phaphyphila sp.Alg1 strain:
[0048] Defluviitalea phaphyphila sp.Alg1[Ji S Q,Wang B,Lu M,etal. Defluviitalea phaphyphila sp.nov.,a novel thermophilic bacterium that degrades brown algae[J].Applied and environmental microbiology,2016,82(3 ):868-877.] inoculated into liquid medium BMS, cultured in an anaerobic test tube at 60°C for 2 days; took 10 mL of the cultured bacteria solution, centrifuged at 12,000×g for 5 min, and collected the bacterial precipitate; buffered with TE Wash twice with liquid to remove the medium residue, collect the bacteria by centrifugation, and extract the genome of the bacteria with the bacterial genome DNA extraction kit according to the method of the TIANGEN Bacterial Genome Extraction Kit. Finally, the DNA sample was dissolved in sterile deionized water at 60°C to obtain genomic DNA.
[0049] The components of the above-mentioned liquid medium BMS are as f...
Embodiment 2
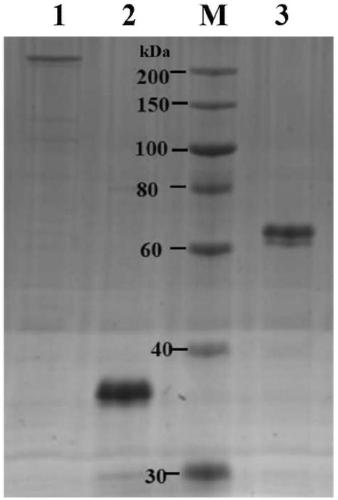
[0055] Cloning of alginate lyase gene and its expression and purification in Escherichia coli strain BL21(DE3):
[0056] 1) According to the genes of three alginate lyases AlgAT0, AlgAT1, AlgAT5, design the following primers:
[0057] AlgAT0:
[0058] Forward primer F: 5'-ATGAATGTTTACGCTACTTCTACTGAAAC-3';
[0059] Reverse primer R: 5'-ATTATCTATACCTACATCTGACGAAGTTAAAGG-3';
[0060] AlgAT1:
[0061] Forward primer F: 5'-GCAAACTATGAAACTTATGATGGTTTTAAAGTT-3';
[0062] Reverse primer R: 5'-TTGTATTGGAAGTAATACTGGTCCTGCTGGATT-3';
[0063] AlgAT5:
[0064] Forward primer F: 5'-CGGAATTCATGAAGGGAAGATTAAAAAAAATGGT-3' (EcoR I);
[0065] Reverse primer R: 5'-CCGCTCGAGACTATGGGTTACTACTAGATTATAAATTTC-3' (Xho I);
[0066] What is underlined in the above-mentioned forward primer is the restriction endonuclease EcoR I site, and what the reverse primer is underlined is the restriction endonuclease Xho I site. The bases shown in bold are protected bases;
[0067] Using the genomic DNA obta...
Embodiment 3
[0167] Embodiment 3, analysis of enzymatic characteristics of alginate lyase:
[0168] a. Substrate specificity assay of alginate lyase
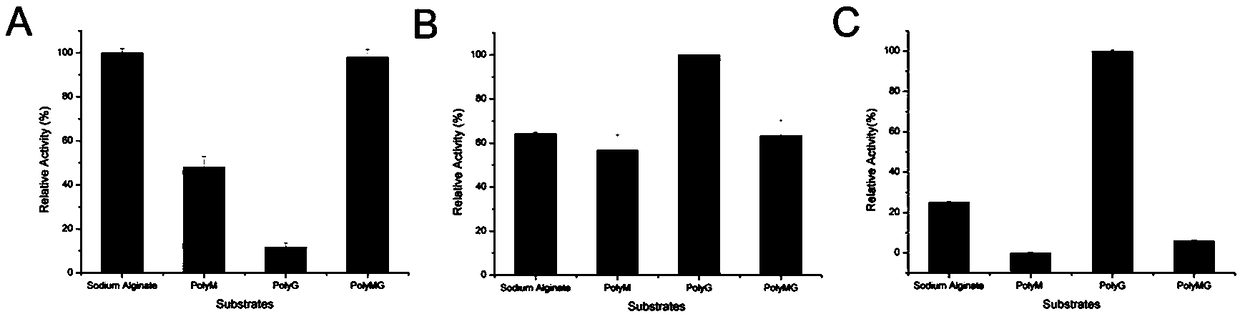
[0169] Purify the above-mentioned examples to obtain alginate lyase AlgAT0, AlgAT1, and AlgAT5 proteins, respectively take 50 μl of enzymes with a concentration of 2 μg / ml, and add them to acetic acid-sodium acetate containing 2 g / L of different substrates (PolyM, PolyG, PolyMG) In the buffer solution (200mM acetic acid-acetic acid sodium salt buffer solution, pH5.8), react at 70°C for 3 minutes, and measure the change value of its OD235nm under the ultraviolet spectrophotometer with water bath circulating heating (see image 3 ). One enzyme activity unit is defined as the value change of 0.1 value per minute of OD235nm. The specific enzyme activity is defined as the ratio of the enzyme activity to the corresponding protein amount.
[0170] Depend on image 3 The results show that AlgAT0 has a specific enzyme activity of 3592U / mg to sodi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Theoretical molecular weight | aaaaa | aaaaa |
| Theoretical molecular weight | aaaaa | aaaaa |
| Theoretical molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com