Recombinant lipase mutant, engineering bacteria and application of recombinant lipase mutant
A technology of mutant and lipase, which is applied in the field of preparation of dimethyl N-acetyl-piperidine-2,3-dicarboxylate, can solve the problem of low concentration of substrate in resolution reaction and low catalytic activity of lipase , long reaction time and other problems, to achieve the effect of less catalyst consumption, short reaction time and high substrate concentration
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
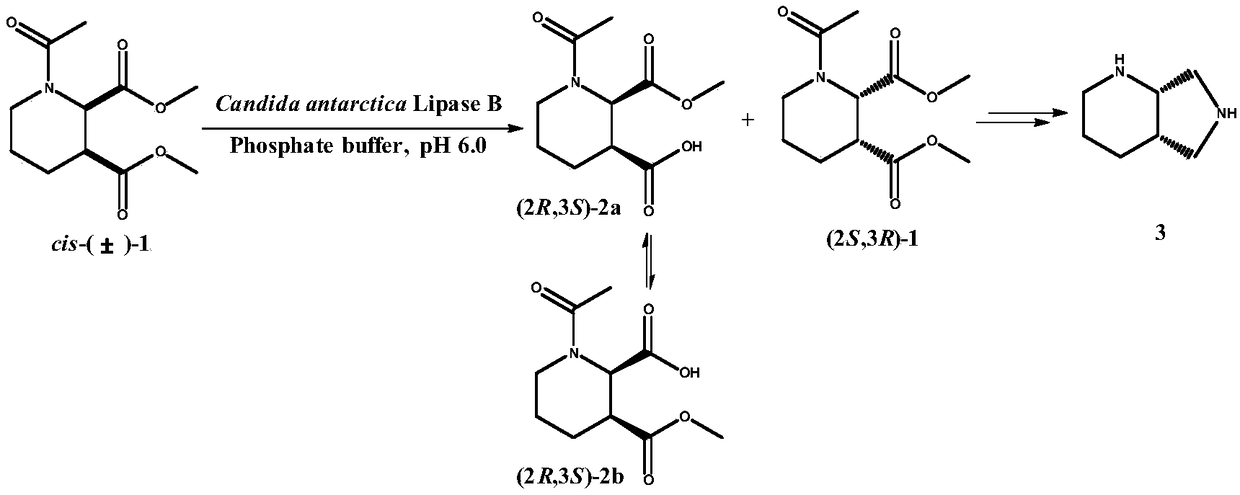
[0026] Embodiment 1: Construction of recombinant lipase genetically engineered bacteria E.coli Rosetta(DE3) / pET22b-CALB
[0027] Candida antarctica lipase B (CALB) gene sequence was optimized by yeast codons to obtain pGEM-T-CALB plasmid by total gene synthesis. Design expression primer 1 (GG CCATGG CCTTACCTAGTGGTTCCGACCCTG), primer 2 (GG GCGG CCG CTCAATGATGATGATGGTGGTGAGGAGTAACAATTCCTGAAC) (the underlines are Nco I and Not I restriction enzyme sites), utilize high-fidelity Pfu DNA polymerase to amplify, obtain the lipase gene sequence of 951bp (nucleotide sequence is as shown in SEQ IN NO.1, The amino acid sequence is shown in SEQ IN NO.2). The amplified fragment was digested with Nco I and Not I restriction endonucleases, and the fragment was ligated with pET22b treated with the same restriction endonucleases using T4 DNA ligase to construct the expression vector pET22b-CALB. The constructed expression vector was transformed into E.coli Rosetta(DE3) competent cells to...
Embodiment 2
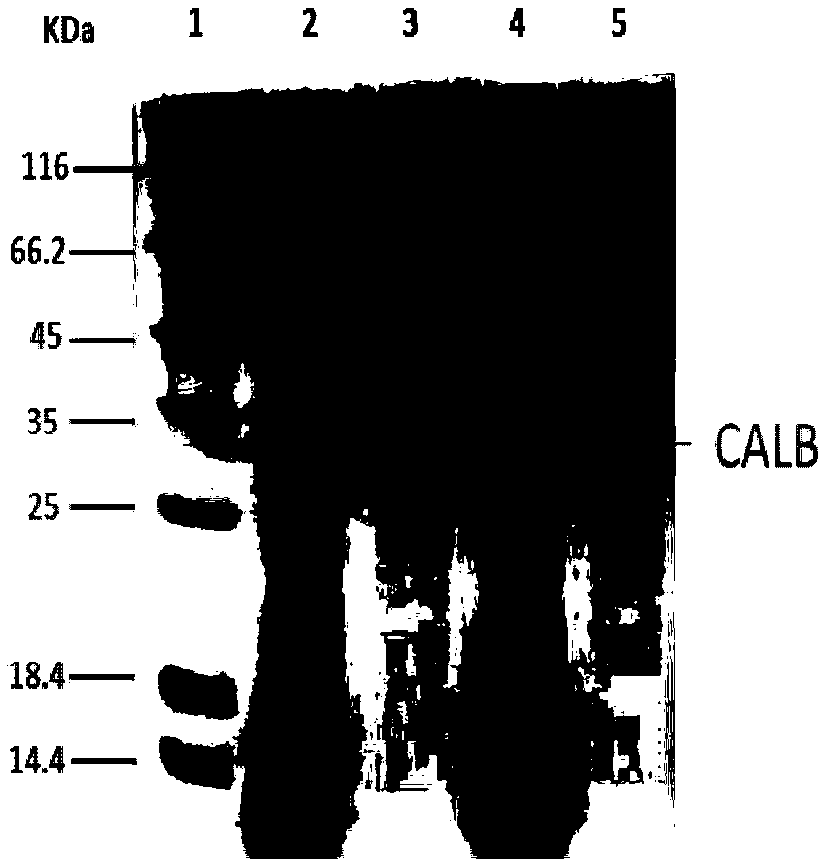
[0028] Example 2: Expression and screening of recombinant lipase mutants
[0029] Using the recombinant bacteria (E.coli Rosetta(DE3) / pET22b-CALB) containing the expression vector pET22b-CALB as the starting strain, the site-directed saturation mutation technique was used to further improve the ability of lipase to the substrate racemic N-acetyl-piperidine- Catalytic activity of dimethyl 2,3-dicarboxylate. Primers were designed as follows:
[0030] Leu / Ala(L / A)140 / 141 combination:
[0031] Upstream primer 5: 5'-ACTACAAAGGTACCGTGNDTNDTGGTCCACTTGACGCCTT-3'
[0032] Downstream primer 6: 5'-GAGAACTGTGATGTCAAAAGATCCTGCATGATCGATAAC-3'
[0033] Leu(L)144:
[0034] Upstream primer 7: 5'-AGGTACCGTGTTGGCTGGTCCANNKGACGCCTTGGCAGT-3'
[0035] Downstream primer 8: 5'-GGACACTGCCAAGGCGTCMNNTGGACCAGCCAACACGGT-3'
[0036] Ile (I) 189:
[0037] Upstream primer 9: 5'-CTCAGCTACAGACGAA NNK GTTCAGCCTCAAGTTAGT-3'
[0038] Downstream primer 10: 5'-CTAACTTGAGGCTGAAC MNN TTCGTCTGTAGCTGAG-3' ...
Embodiment 3
[0050] Embodiment 3: Construction of wild-type and mutant recombinant lipase genetically engineered bacteria Pichia pastoris X-33 / CALB and Pichia pastoris X-33 / CALB-muts
[0051] Using the pGEM-T-CALB plasmid as a PCR template, the expression primer 3 (GG CTCGAG AAAAGAGAGGCTGAAGCTTTACCTAGTGGTTCCGACC), primer 4 (GGT CTAGA TCAATGATGATGATGGTGGTGAGGAGTAACAATTCCTGAA) (the underlines are Xho I and Xba I restriction enzyme sites), utilize high-fidelity Pfu DNA polymerase to amplify, obtain the lipase gene sequence of 951bp (nucleotide sequence as shown in SEQ ID NO.1) . Use Xho I and Xba I restriction endonucleases to digest the amplified fragment, use T4 DNA ligase to connect the fragment with pPiczα-A treated with the same restriction endonuclease, and construct the vector pPiczα-A-CALB . The constructed vector was linearized with Sca I, and the linearized pPiczα-A-CALB was introduced into Pichia pastoris X-33 by electric shock transformation method, and then integrated into t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com