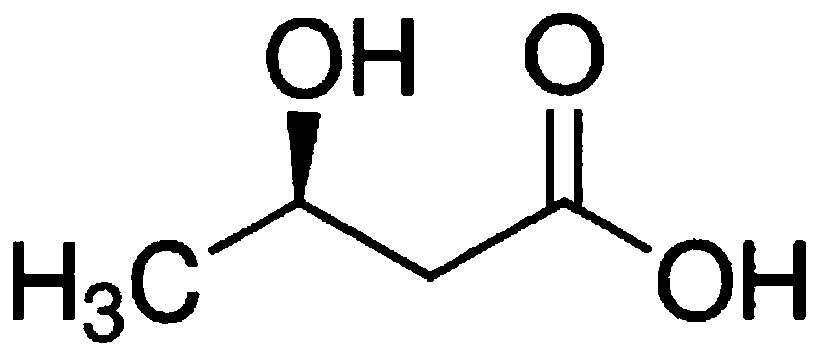
Recombinant ketoreductase and application thereof to preparation of (R)-3-hydroxybutyric acid and salt thereof
An R-3-, hydroxybutyric acid technology, applied in the field of recombinant protein, can solve the problems of difficult separation and purification, difficult industrial application, low fermentation unit, etc., and achieve breakthroughs in low chiral purity, low production cost, and high cost reduction. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] (1) Wild CmCR Ketoreductase Engineering Bacteria
[0072] The ketoreductase shown in SEQ ID NO: 1 (GenBank: AB036927, derived from the nucleotide sequence of Candida magnoliae) was synthesized through the whole gene sequence, and then connected with the plasmid pET28a (commercially available) to obtain the recombinant The vector pET28a-CmCR; then the recombinant vector was introduced into Escherichia coli BL21(DE3) to construct the Escherichia coli genetic engineering strain pET28a-CmCR / BL21(DE3) expressing ketoreductase.
[0073] The amino acid sequence of wild ketoreductase is shown below:
[0074] MAKNFSNVEYPAPPPAHTKNESLQVLDLFKLNGKVASITGSSSGIGYALAEAFAQVGADVAIWYNSHDATGKAEALAKKYGVKVKAYKANVSSSDAVKQTIEQQIKDFGHLDIVVANAGIPWTKGAYIDQDDDKHFDQVVDVDLKGVGYVAKHAGRHFRERFEKEGKKGALVFTASMSGHIVNVPQFQATYNAAKAGVRHFAKSLAVEFAPFARVNSVSPGYINTEISDFVPQETQNKWWSLVPLGRGGETAELVGAYLFLASDAGSYATGTDIIVDGGYTLP
[0075] The nucleotide sequence of wild ketoreductase is as follows:
[0076] ATGGCTAAGAA...
Embodiment 2
[0084] Example 2 Construction of Ketoreductase Saturation Mutant Library
[0085] (1) Computer simulation of CmCR ketoreductase structure modeling
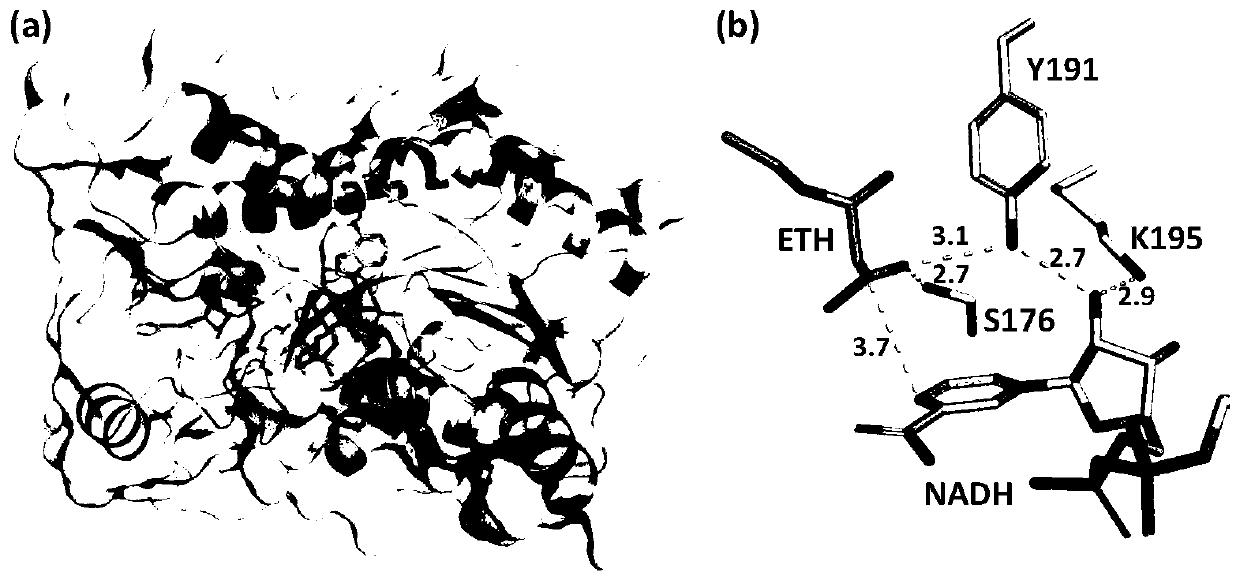
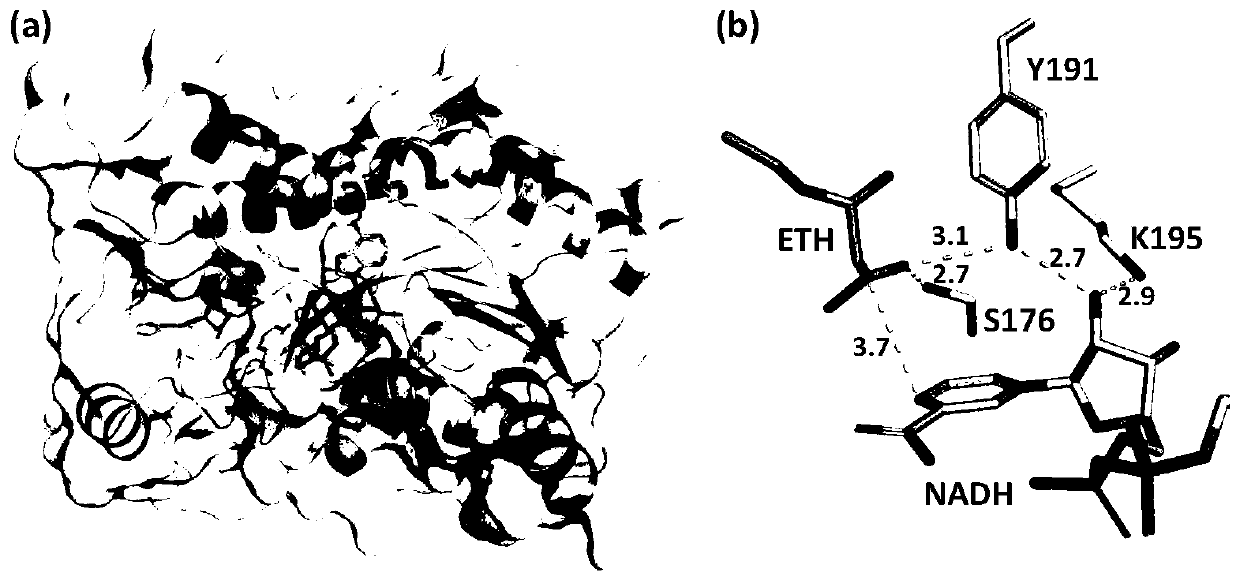
[0086] CmCR derived from Candida magnoliae is homologous to carbonyl reductase (PDB: 3CTM) (CACR for short) derived from Candida parapsilosis. Homologous modeling was carried out by Sybyl software to obtain its 3D structure ( figure 2 a). According to the 3D structure diagram of homology modeling, it was found that the three sites S176, Y191, and K195 in CmCR played an important role in the reaction with ethyl acetoacetate as the substrate ( figure 2 b).
[0087] (2) Molecular Construction of Saturation Mutation Library
[0088] According to the computer three-dimensional structure simulation results, S176, Y191, and K195 were subjected to saturation combination mutations to establish a saturation mutation gene bank. The specific method is as follows:
[0089] ①Primer design
[0090] Using the sequence of SEQ ID NO: 1 as a ...
Embodiment 3
[0125] The fermenter fermentation of embodiment 3 recombinant strains
[0126] (1) Recombinant ketoreductase mutant strain CmCR-186 / BL21 (DE3) 7 liter fermenter fermentation
[0127] Inoculate the single clone of the recombinant ketoreductase mutant strain CmCR-186 / BL21(DE3) into 4 mL of LB medium containing 50 mg / L kanamycin resistance, culture at 37°C and 200 rpm for 20 h, and then take 2 mL of the overnight culture The cells were transferred to 200mL seed medium, and after culturing at 37°C and 200rpm for 4h, all the 200mL cultured cells were inoculated into a 7L fermenter containing 4.8L fermentation medium for fermentation. Fermentation tank culture conditions: tank temperature 37°C, aeration ratio 2.5vvm, stirring 350rpm, tank pressure 0.05MPa, when the OD600 of the bacteria in the fermentation broth = 5-6, add ITPG with a final concentration of 0.2mM for induction, and lower the temperature to 25°C , the tank pressure, air flow, etc. remain unchanged, continue to culti...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
| specific rotation | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com