A High Coverage Lipidomics Analysis Method Based on Liquid Chromatography-Mass Spectrometry
A lipid group, high-coverage technology, applied in the fields of analytical chemistry, biochemistry and medicine, can solve problems such as poor linear response of mass spectrometry, influence of quantitative accuracy, and difficult data repetition, so as to simplify the data processing process and improve repeatability , the effect of accurate quantitative ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 High coverage lipidomics analysis method based on UPLC / QQQ-MS-DMRM
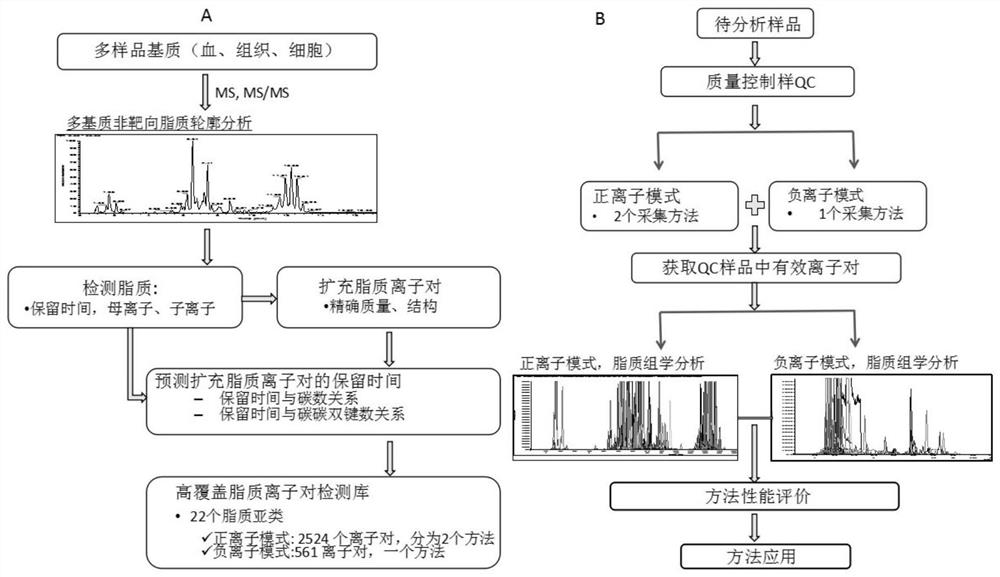
[0025] The flowchart of the establishment of the high-coverage lipidomics analysis method based on UPLC / QQQ-MS-DMRM is as follows figure 1 Shown in A, the specific implementation steps are as follows:
[0026] 1. Preparation of different types of biological samples.
[0027] Eight exogenous metabolites were added to the extraction solutions of lipid metabolites in different types of samples as internal standards, internal standard mixture PC38:0, 6.7μg / ml; LPC19:0, 3.3μg / ml; TG45:0, 5.3 μg / ml; PE34:0 / PE30:0, 3.3μg / ml; FFA16:0-d3, 6.7μg / ml; FFA18:0-d3, 6.7μg / ml; Cer35:1, 1.7μg / ml; SM30: 1, 1.7 μg / ml.
[0028] 1) Tissue samples: operate on ice, weigh 10mg rat brain tissue into a 2ml EP tube, add grinding beads, add 30μl internal standard mixture, add 400μl methanol solution, grind for 25HZ*1min*2times, add 800μl chloroform , shake for 10 minutes, add 240 μl of ultra-pure water, shake for 5 ...
Embodiment 2
[0044] Example 2. Analysis of plasma high coverage lipidomics
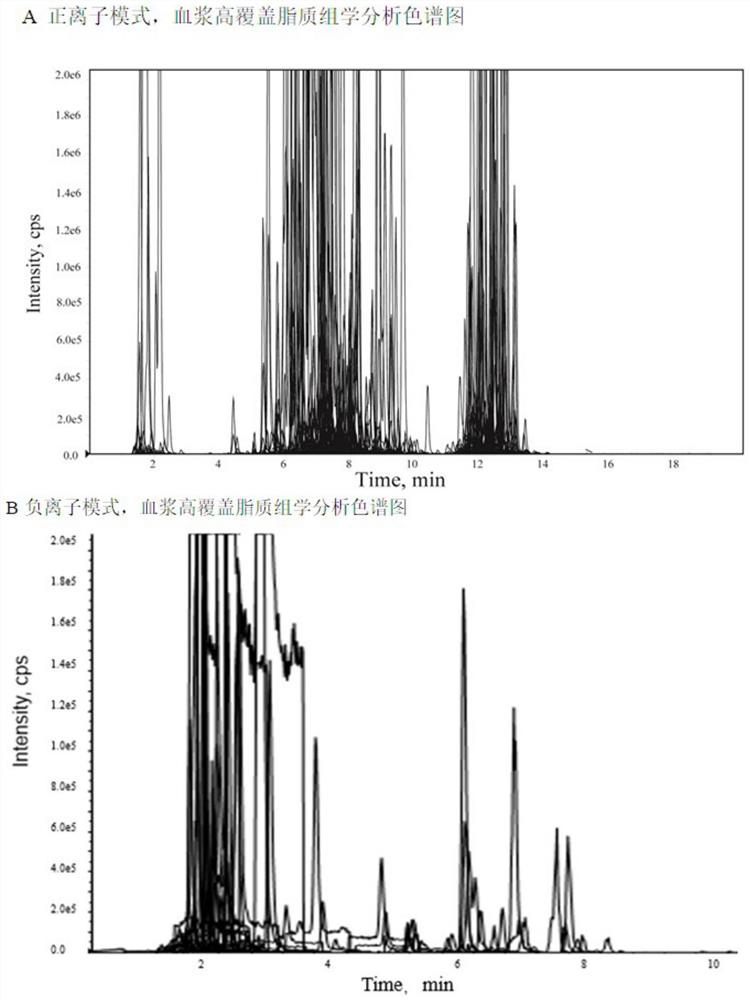
[0045] The specific process of plasma high-coverage lipidomics analysis is as follows: figure 1 Shown in B, concrete implementation steps are as follows:
[0046] 1. Plasma sample pretreatment
[0047]First, mix the plasma samples to be tested in equal amounts, as the quality control sample QC, take 40 μl QC, add 300 μl methanol solution (containing 8 internal standards, PC38:0, 0.67 μg / ml; LPC19:0, 0.33 μg / ml; TG45: 0, 0.53μg / ml; PE34:0 / PE30:0, 0.33μg / ml; FFA16:0-d3, 0.67μg / ml; FFA18:0-d3, 0.67μg / ml; Cer35:1, 0.17μg / ml ;SM30:1, 0.17μg / ml), vortex for 10s, add 1ml MTBE, shake for 10min, add 300μl ultrapure water, vortex for 30s, stand at 4℃ for 10min, centrifuge at 10000rpm*4℃*10min, take the upper layer 400μl frozen Dry, redissolve (reconstituted solution: dichloromethane / methanol=2:1; diluent: acetonitrile / isopropanol / water=65:30:5; reconstituted solution / diluted=1:3, V / V) to 120μl, used for mass spectrometr...
Embodiment 3
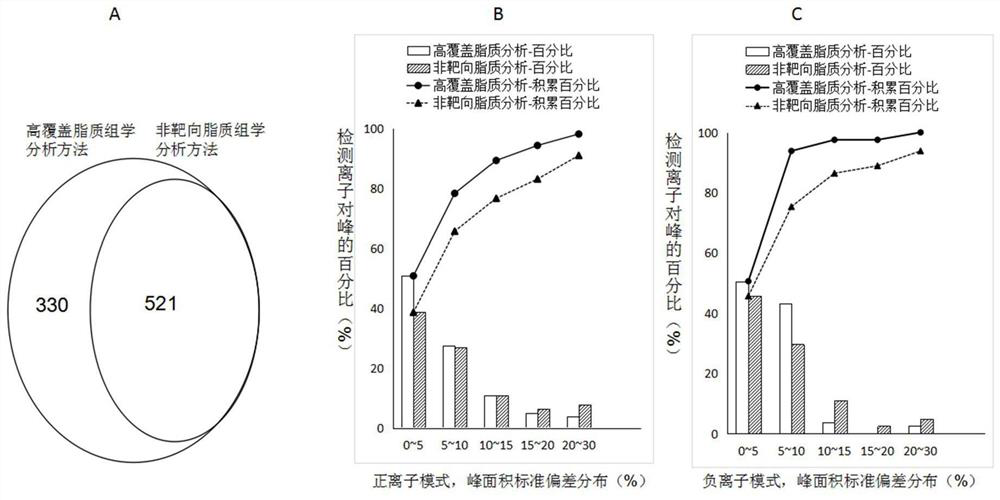
[0050] Example 3. Methodological investigation of high-coverage lipidomics analysis based on UPLC / QQQ-MS-DMRM
[0051] The above-mentioned plasma samples to be tested were mixed in equal volumes to prepare a quality control sample (QC), which was pretreated according to the following different investigation objects.
[0052] 1. Repeated inspection
[0053] Pipette 40μl QC, add 300μl methanol solution (containing 8 internal standards, PC38:0, 0.67μg / ml; LPC19:0, 0.33μg / ml; TG45:0, 0.53μg / ml; PE34:0 / PE30:0 , 0.33μg / ml; FFA16:0-d3, 0.67μg / ml; FFA18:0-d3, 0.67μg / ml; Cer35:1, 0.17μg / ml; SM30:1, 0.17μg / ml), vortex 10s , add 1ml MTBE, shake for 10min, add 300μl ultrapure water, vortex for 30s, stand at 4℃ for 10min, centrifuge at 10000g*4℃*10min, take 400μl of the upper layer to freeze-dry. Redissolve (reconstituted solution: dichloromethane / methanol=2:1; diluent: acetonitrile / isopropanol / water=65:30:5; reconstituted solution / diluted=1:3, V / V) to 200 μl, For negative ion analysis,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com