Plasmid, phage-assisted continuous directed evolution system and directed evolution method
A directed evolution, bacteriophage technology, applied in biochemical equipment and methods, microorganism-based methods, bacteria, etc., can solve problems such as being unsuitable for directed evolution of membrane proteins and not involving the field of membrane proteins.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
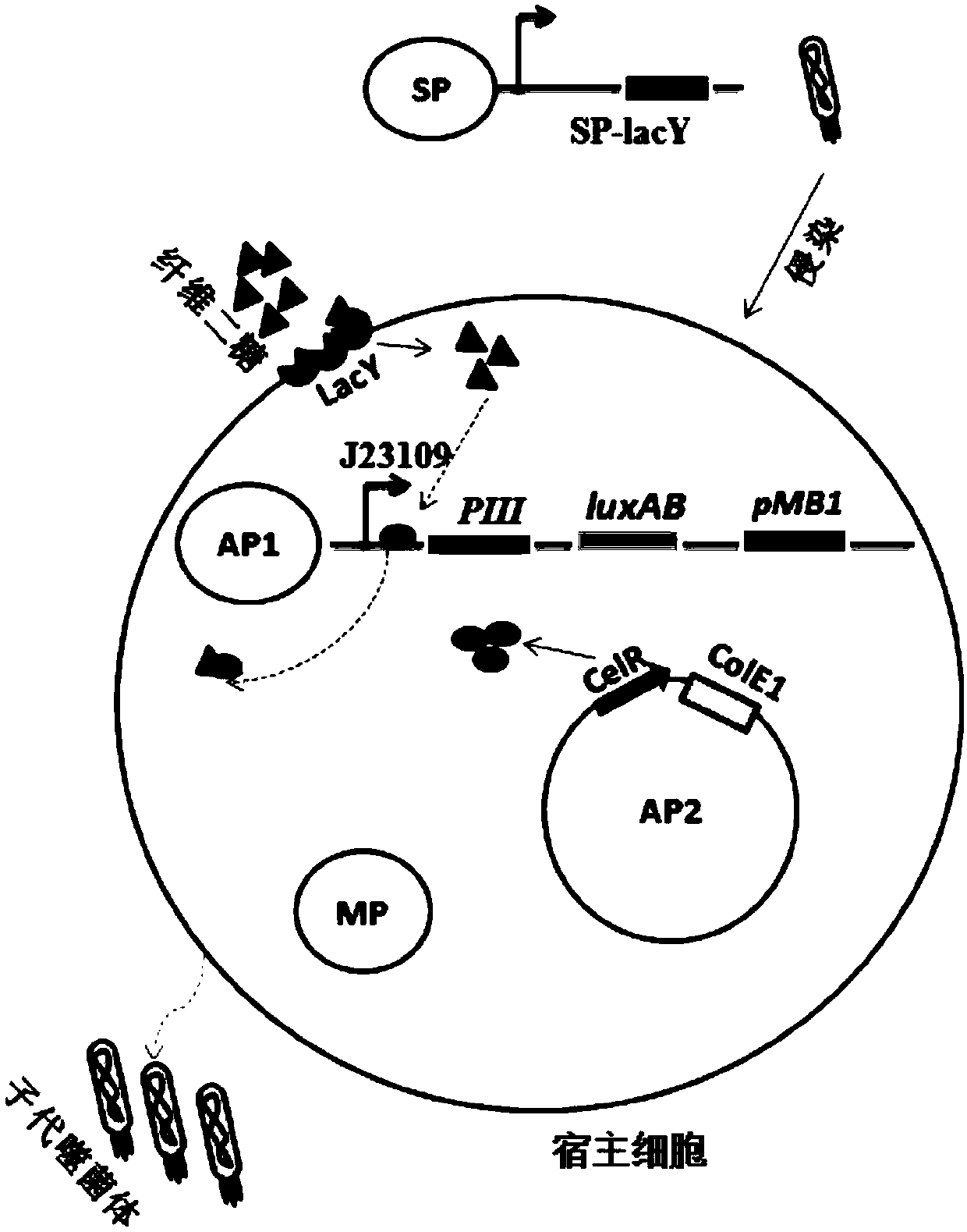
[0055] Packaging of bacteriophage SP-lacY carrying quasi-evolved genes
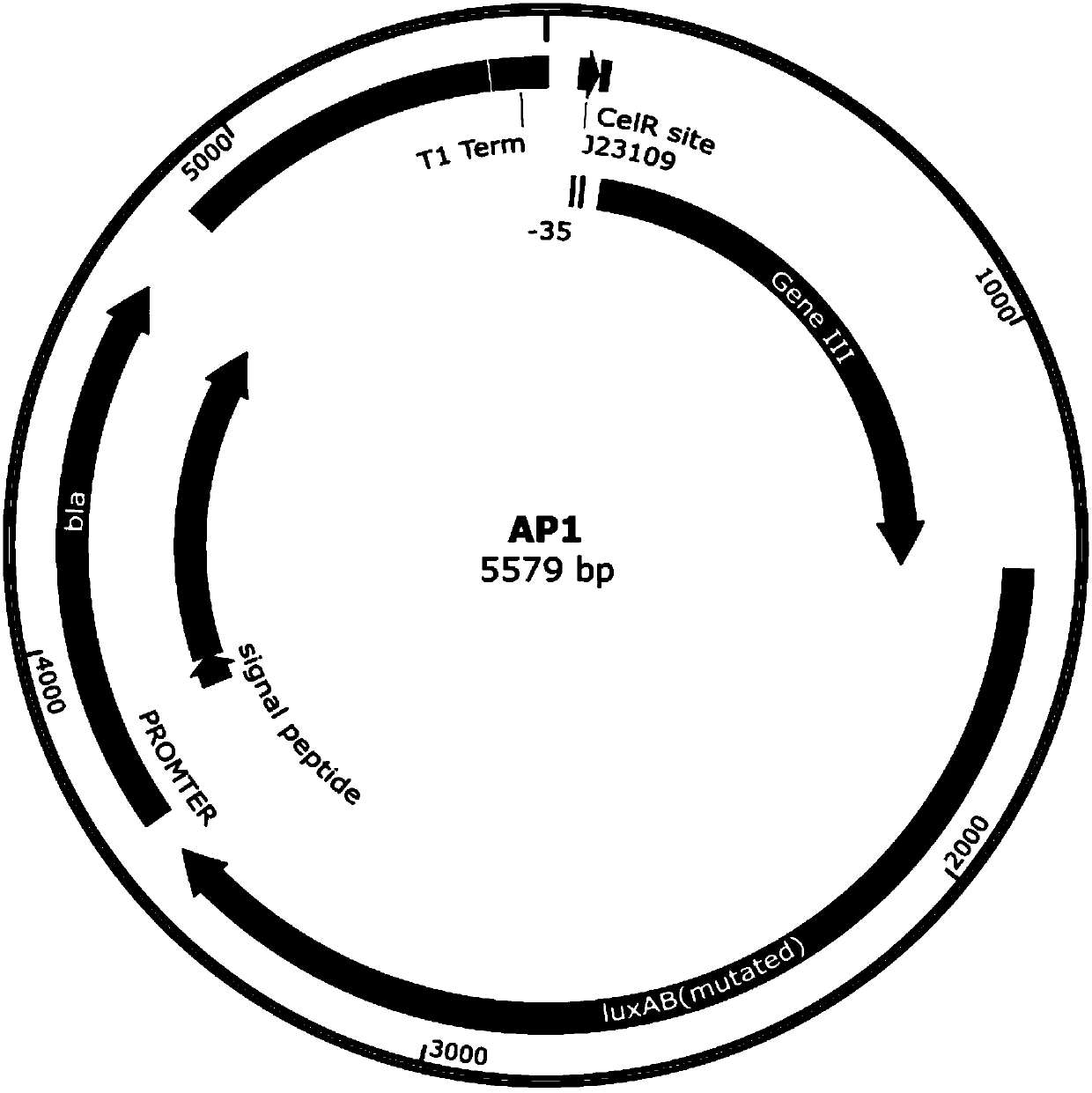
[0056] 1) Construction of the gIII protein expression plasmid AP1: the gIII protein is promoted by the phage shock promoter J23109, and the nucleic acid sequence (SEQ ID NO.3) of the recognition site of the CelR protein is inserted downstream of the promoter. AP1 map see figure 2 .
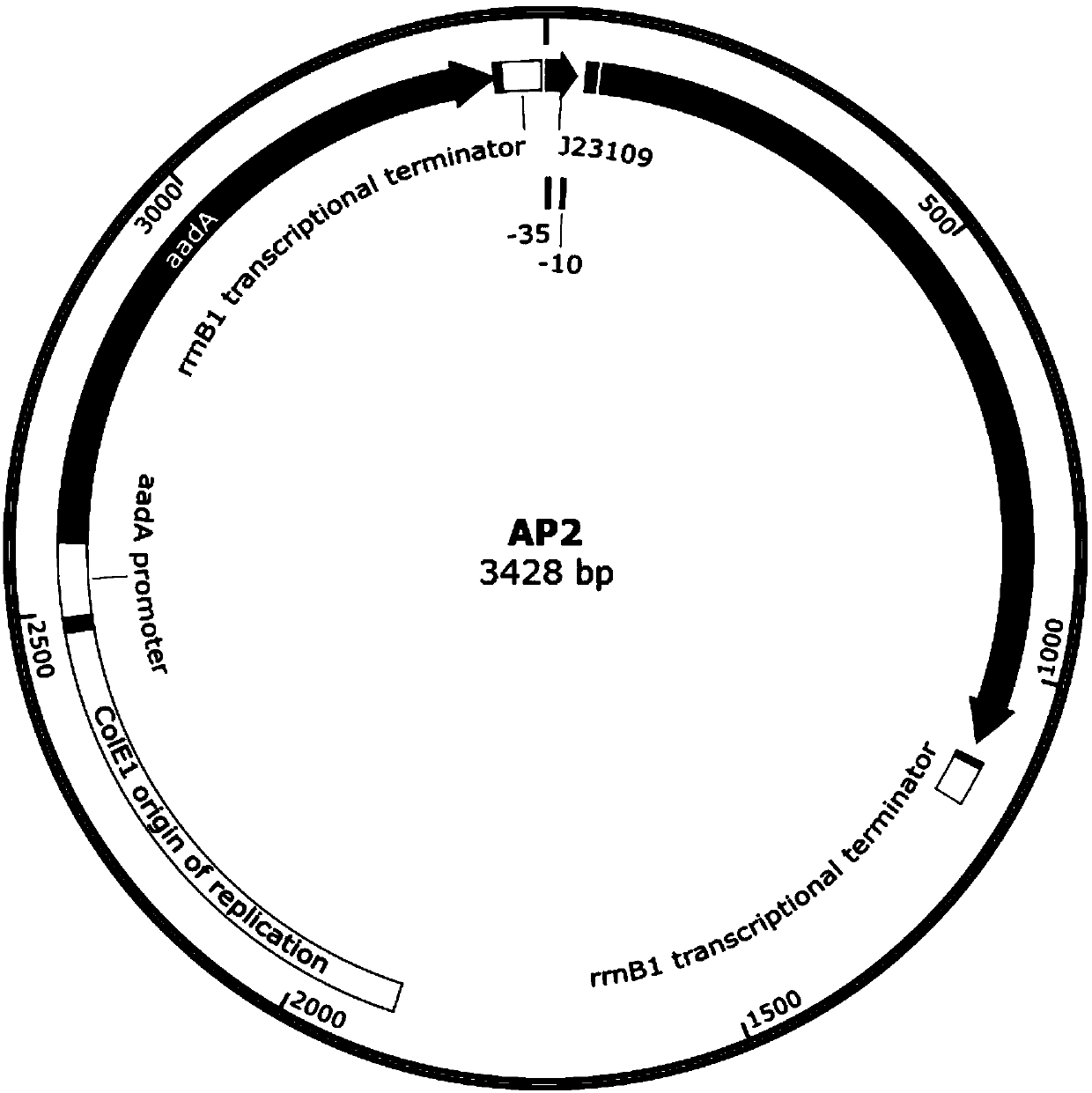
[0057] 2) Construction of expression plasmid AP2 of functional protein CelR protein: CelR is expressed by a constitutive promoter ( image 3 ).
[0058] 3) The mutagenic plasmid MP was donated by David R Liu's laboratory, see the map Figure 4 .
[0059] 4) AP1, AP2 and MP co-transform S1030 competent cells to obtain host S1030-AP1 / AP2 / MP. Before the host does not have LacY protein to transport cellobiose into the cell, the CelR protein expressed on AP2 and the CelR protein on AP1 The CelR recognition site binds to repress the expression of the downstream gene gIII; MP is a plasmid that improves mutations and is induced...
Embodiment 2
[0065] Verification of the relationship between the concentration of cellobiose and the propagation speed of bacteriophage SP-lacY
[0066] 1) The host S1030-AP1 / AP2 / MP LB medium was cultured to the logarithmic phase (OD 600 = 0.4).
[0067] 2) Dilute the wild-type SP-lacY phage and add it to the above logarithmic phase host to make the initial concentration in the system 50pfu / mL.
[0068] 3) The final concentration gradient of cellobiose is 29mM, 14.5mM, 2.9mM, 1.45mM, 0.29mM, 0.0029mM, 0.00029mM, 0.00mM.
[0069] 4) Take samples every 15 minutes for each cellobiose gradient, and detect the proliferation of phage SP-lacY in the system. The results are as follows: Image 6 shown.
[0070] Image 6 In , the concentration of cellobiose from 0mM to 0.029mM basically coincides with the abscissa.
[0071] Image 6 The experimental results show that the proliferation rate of phage is directly proportional to the concentration of cellobiose, and the reduction of the concentrat...
Embodiment 3
[0073] Substrate specificity of PACE evolved LacY
[0074] 1) The host S1030-AP1 / AP2 / MP was cultured to the logarithmic phase (OD 600 = 0.4).
[0075] 2) The first round of evolution, in the 1mL evolution system, the initial cellobiose final concentration was 29mM, and the initial wild-type SP-lacY phage was 1.2×10 5 pfu / mL, the final concentration of arabinose is always 1%, add host S1030-AP1 / AP2 / MP to 1mL, 37°C, 150RPM for 1h. Sampling, serial dilution, mixed 10 μL of diluted phage with 190 μL logarithmic phase host S1030-AP1 / AP2 / MP, placed at 37°C for 15 minutes, mixed with 1mL of 0.5% soft agar containing 0.5% cellobiose at 50°C, and evenly Spread on a solid agar plate with a diameter of 60 cm, let it stand for 10 minutes, and culture it overnight at 37°C after solidification, calculate the number of phage plaques, and determine the concentration of phage in the system. Take a single plaque, use primers SP1-F and SP1-R to amplify the LacY gene fragment on the phage, and...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Titer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com