Argonaute protein mutant and application thereof
A mutant and protein technology, applied in the field of Argonaute protein mutants and their applications, can solve the problems of time-consuming, limited range of target DNA, complicated operation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Embodiment 1: Preparation of the Ago protein mutant of the present invention
[0086] Step 1: Construction of expression vector
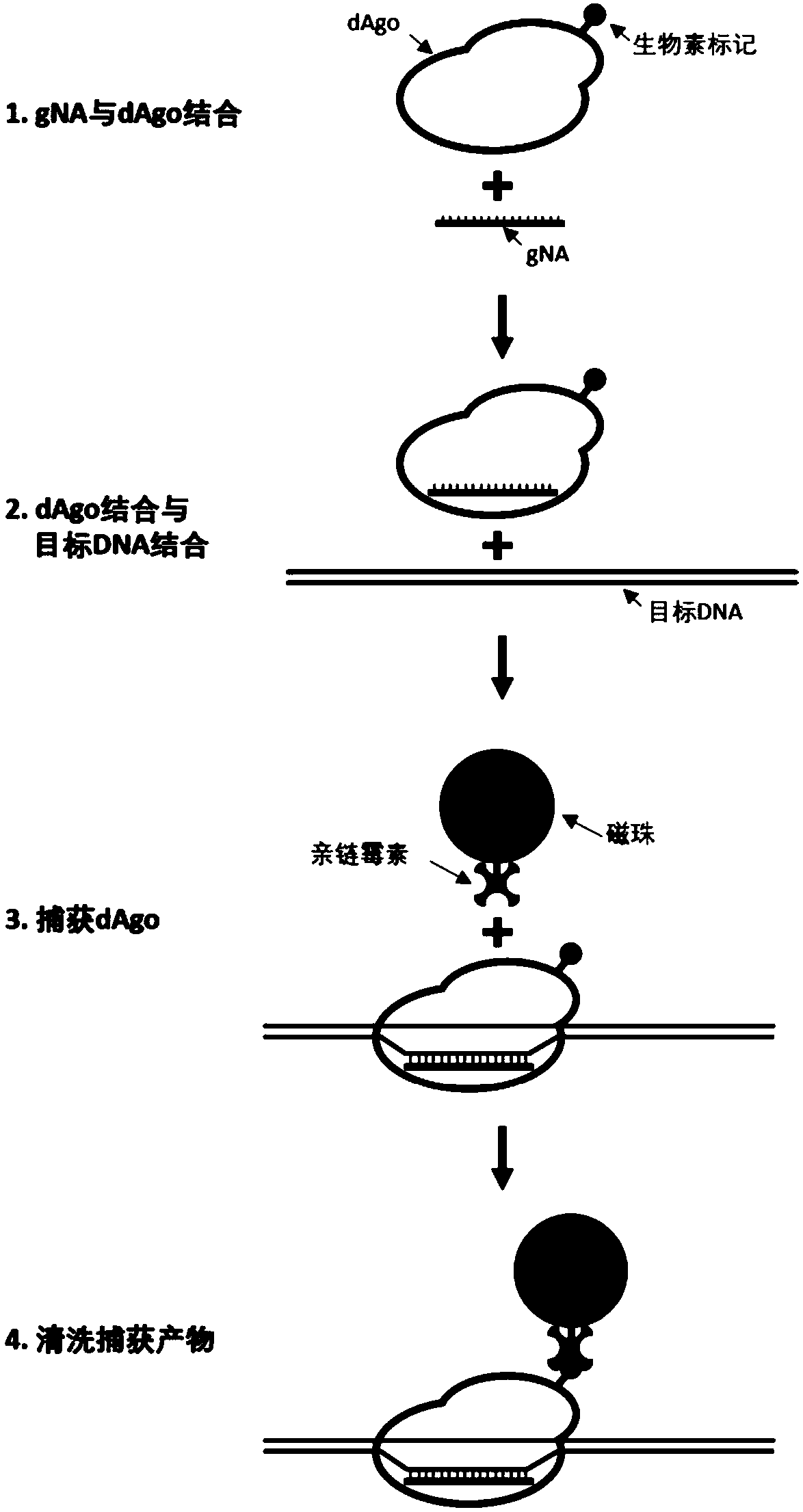
[0087] A biotin receptor sequence was connected to the N-terminal of the known amino acid sequence (SEQ ID NO: 1) of the Pyrococcus furiosus Ago protein (PfAgo), and a codon-optimized codon for Escherichia coli (E.coli) was designed and synthesized accordingly the nucleotide sequence. The nucleotide sequence, 6x His-Tag, PfAgo-BAS, IRES, BirA (Escherichia coli biotin ligase) were sequentially cloned into the pET-28a vector with the kanamycin resistance gene to obtain the vector pPFA-1.0.
[0088] Using the Q5Site-Directed Mutagenesis Kit (NEB, Cat#E0554S), the pPFA-1.0 was subjected to site-directed mutagenesis according to the operating procedures in the manual. The DNA obtained after mutation was transformed into E. Coli DH5α cells, and cultured overnight at 37°C in LB agarose medium containing kanamycin. For each mutation, 10 colonie...
Embodiment 2
[0100] Embodiment 2: enrich target DNA according to the method of the present invention
[0101] The target DNA in this example is the exon 18-21 fragment of EGFR gene from free DNA in plasma samples and genomic DNA in leukocytes isolated from normal human peripheral blood, respectively.
[0102] Step 1: Extract DNA
[0103] For free DNA: take 4mL of human plasma, use QIAamp Circulating Nucleic Acid Kit (Qiagen, Cat#55114) to extract free DNA according to the kit instructions, and then elute with 45uL Elution Buffer.
[0104] For genomic DNA: take 200uL of leukocytes isolated from human peripheral blood, use MagJET Whole Blood gDNAKit (ThermoFisher, Cat#K2741), and extract genomic DNA according to the instructions of the kit. About 500 ng (30 uL) of the extracted genomic DNA was sonicated (Sonicator Biorupter Pico from Diagenode SA).
[0105] Step 2: Design Guide DNA (gDNA)
[0106] According to the sequence of EGFR exons 18, 19, 20, and 21, gDNA with 5' phosphorylation m...
Embodiment 3
[0121] Embodiment 3: construct the sequencing library of target DNA according to the method of the present invention
[0122] Step 1: Cell-free DNA extraction
[0123] Take 4mL of human plasma, use QIAamp Circulating Nucleic Acid Kit (Qiagen, Cat#55114) to extract free DNA according to the kit instructions, and finally free DNA is eluted with 45uL Elution Buffer provided by the kit.
[0124] Step 2: Ligation of Sequencing Adapters
[0125] Using the KAPA Hyper Prep Kit (Kapa Biosystems, Cat#KK8501) according to the instructions, the free DNA was filled in at the end and A was added, and then ligated with the TruSeq adapter suitable for the Illumina sequencing platform.
[0126] Step 3: Pre-amplification of Ligation Products
[0127] Prepare the reaction system according to the following table:
[0128]
[0129] On the PCR instrument, perform pre-amplification according to the following conditions:
[0130]
[0131]
[0132] After the amplification was complete, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com