Application of single cell sequencing in immune cell analysis
A single-cell sequencing and immune cell technology, applied in the application field of single-cell sequencing in immune cell analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
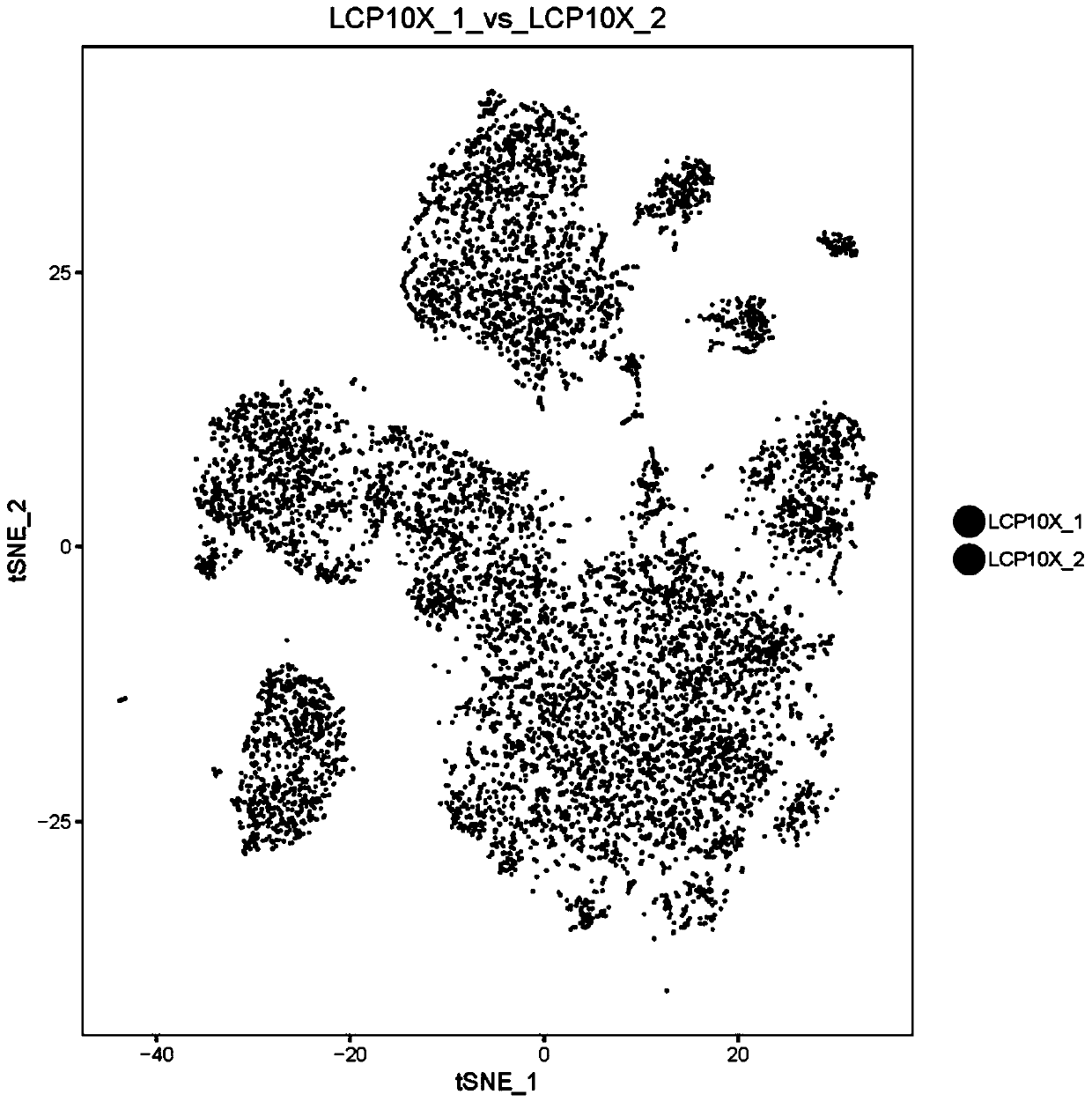
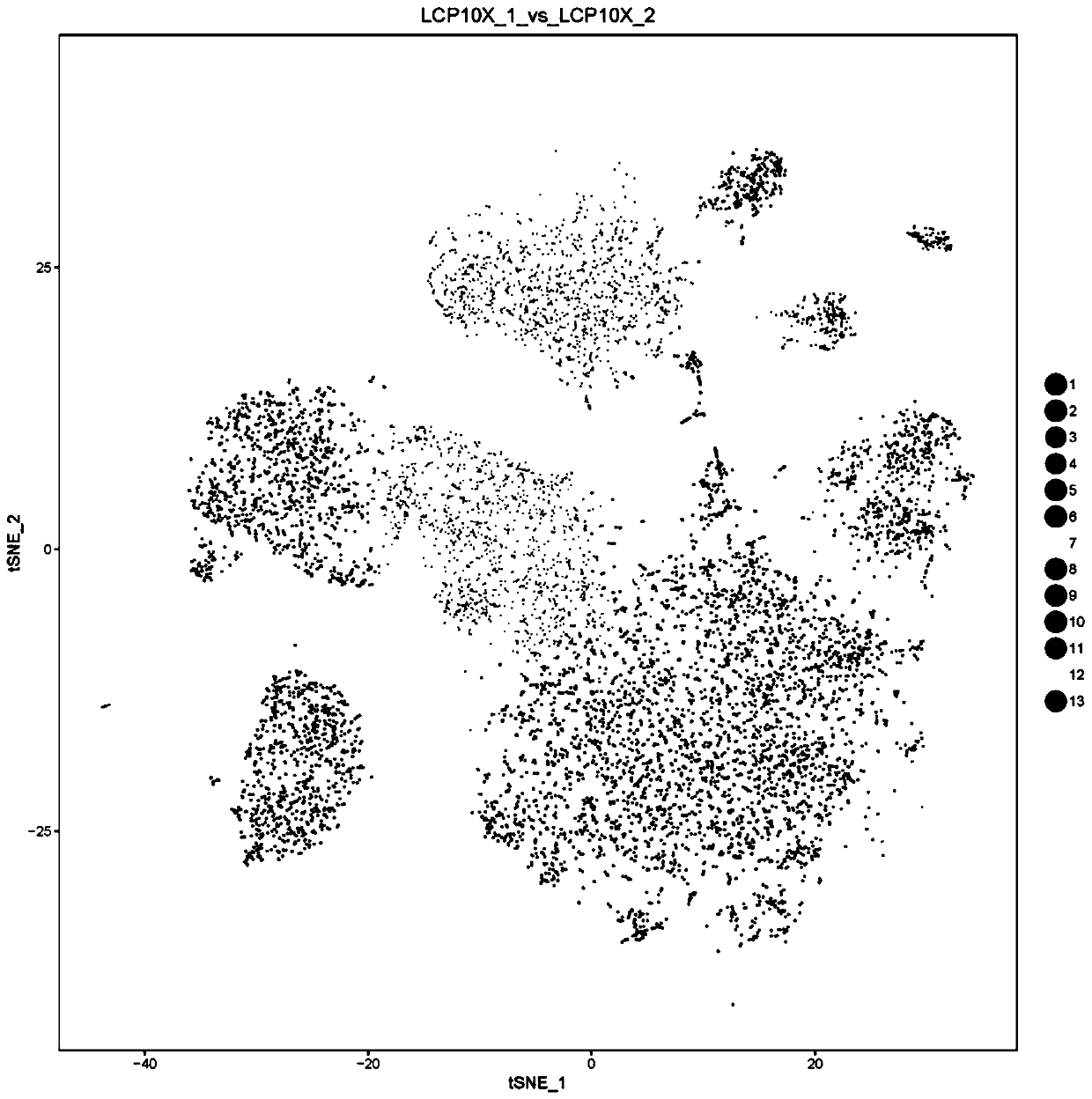
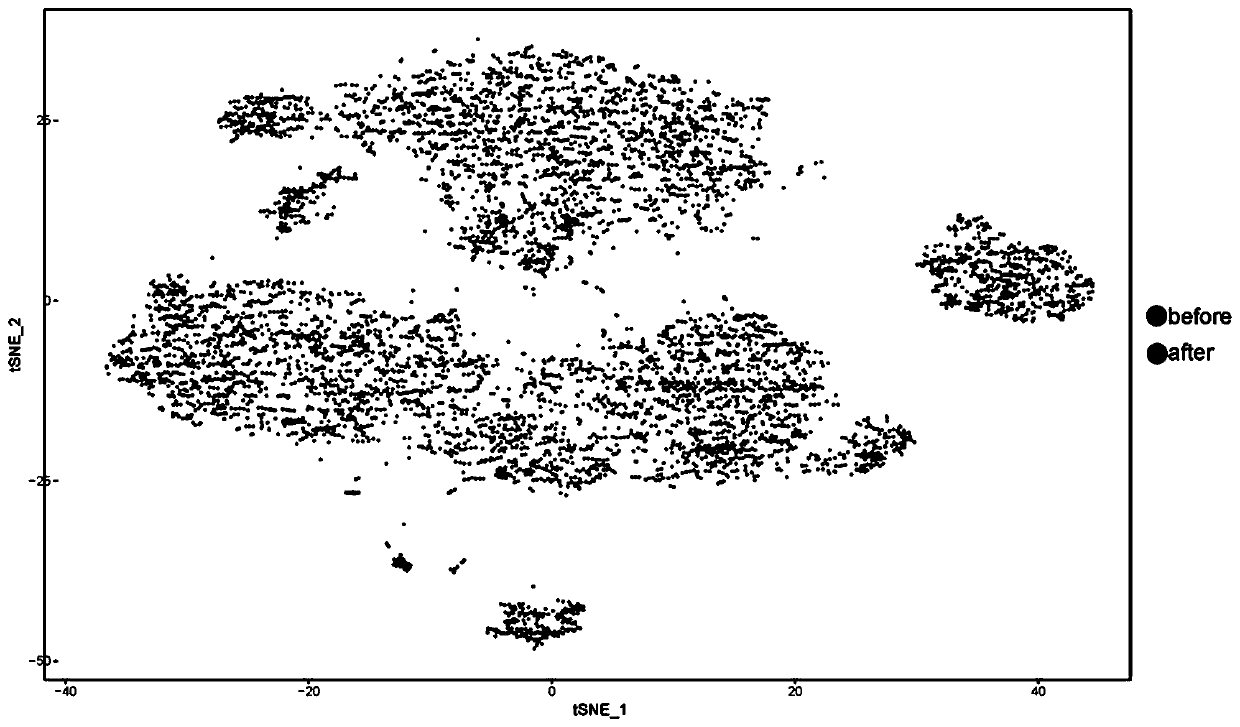
Image
Examples
Embodiment 1
[0046] Example 1 Construction and sequencing of single-cell transcriptome
[0047] (1) Single cell pretreatment
[0048] use Massive Single Cell mRNA Sequencing Preprocessing Kit processes single cells, including the following steps:
[0049] ①Preparation steps:
[0050] Chip preparation: the small hole end of the chip is the sample inlet, and the big hole end is the sample outlet, so that both the big and small holes are filled with 0.02% PBS-Tween liquid, put the chip in a vacuum desiccator for 15 minutes, take it out and put it at room temperature Let it stand for 15 minutes to ensure that there are no air bubbles in the chip;
[0051] Lysis buffer (lysis buffer) and magnetic beads (beads) preparation: prepare the lysate on ice, and wash the magnetic beads with PBS buffer;
[0052] Cell preparation: resuspend the cells in PBS, and adjust the corresponding cell concentration according to the expected number of cells;
[0053] ② Chip operation:
[0054] Inject the resu...
Embodiment 2
[0063] Embodiment 2 biological information analysis
[0064] The sequencing data analysis method of this embodiment includes:
[0065] (1) Barcode quality control analysis
[0066] Before the overall quality control, first perform barcode data quality analysis, filter out the cellbarcode with low sequencing quality to ensure the accuracy of subsequent analysis, filter the number of bases in the barcode with a quality value lower than 14 or greater than 1; in addition, for UMI The subsequent reads without polyA are also filtered;
[0067] In order to obtain high-quality data for subsequent analysis, use fastp to trim Reads2 and filter reads. The filtering content is as follows:
[0068] Truncating the end adapter sequence;
[0069] Remove more than 5 reads containing N (N means that the base information cannot be determined);
[0070] According to the 4-base sliding window from the 5' end to the 3' end of the reads, the 3' end sequence of the window whose average quality va...
Embodiment 3
[0080] Example 3 Cell Subtype Annotation
[0081] Using the semi-automated annotation of cell subgroups independently developed by Xinge, as well as the reference data set and knowledge base in the Xinge database to annotate cell subtypes, the steps are as follows:
[0082] (1) Semi-automatic cell subgroup annotation: According to the average expression level and cell expression percentage of the marker gene of immune cells in each cluster, the cell type of each cluster is obtained, and this work is completed by running a script;
[0083] (2) Manual verification:
[0084] First, understand the sample type (species, tissue / peripheral blood, tumor / adjacent cancer) based on the sample information, and then judge the main cell type of immune cells based on the clinicopathological information related to the sample (stage, curative effect, etc.);
[0085] According to the gene expression in the differentially expressed gene list of each subgroup, evaluate the possible subtypes. The...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com