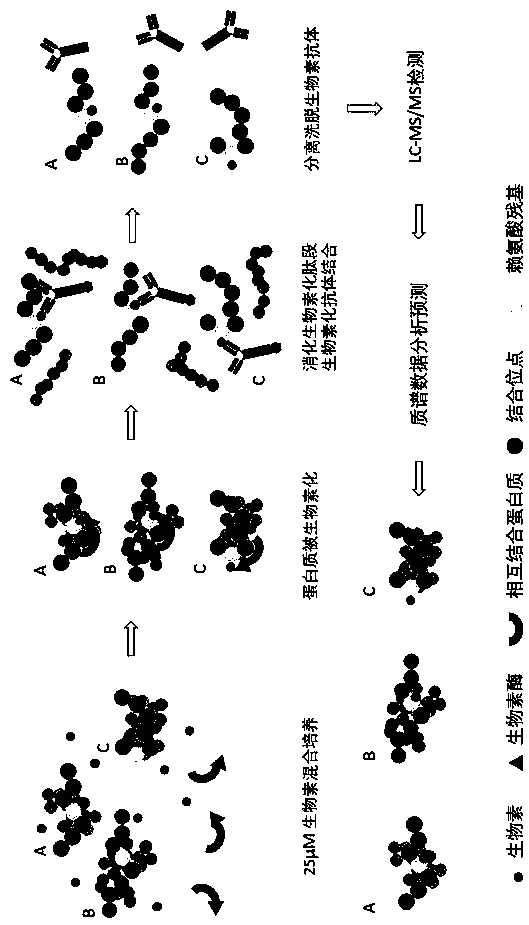
Preparation method of specific peptide fragment mass spectrometry sample
A mass spectrometry, species-specific technology, applied in the field of biochemistry, can solve the problems of ignoring the detection and analysis limitations of mass spectrometry technology, and achieve the effect of low affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Embodiment 1: Preparation method of HEK293T cell-specific peptide mass spectrometry analysis sample:
[0024] A: Take the HEK293T cells to be analyzed, collect 5 million cells in a biotin culture environment, and dissolve them in 2 ml of cell lysis buffer. The composition of the cell lysis buffer is 50 mM triethylammonium bicarbonate, 8 M urea, 10nm protease inhibitor, lysed by circulating ultrasound in an ice bath, quantified the concentration of the protein solution, and then digested the peptide with trypsin;
[0025] B: Trypsin method to digest peptides, a total of 15 mg protein lysis solution, add dithiothreitol at a final concentration of 5 mM, incubate at room temperature for 1 hour, then add iodoacetamide at a final concentration of 10 mM, and incubate at room temperature in the dark For 20 minutes, finally add 3 times the total volume of 50 mM triethylammonium bicarbonate and 150 µg trypsin, and incubate at 37°C with shaking for more than 12 hours; the digested...
Embodiment 2
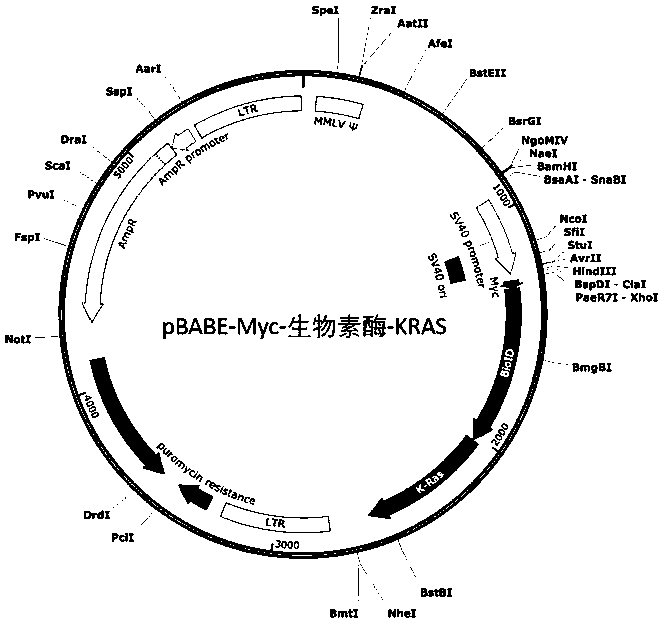
[0033] Example 2: HEK293T cell example of Myc-biotididase-KRAS fusion protein
[0034] (1) Preparation of HEK293T cells expressing Myc-biotidase-KRAS fusion protein
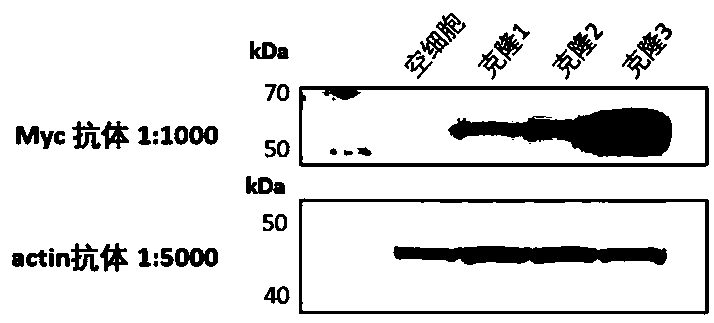
[0035] The gene-synthesized fusion gene fragment containing Myc-tag biotinidase (5' end) and KRAS (3' end) was inserted into the pBABE plasmid behind the SV40 promoter through homologous recombination ( figure 2). The fusion gene fragment plasmid was transiently transfected into HEK293T cells using lipofectamine3000 and packaged into retrovirus. HEK293T cells were infected with virus that was filtered through a 0.22 μm filter to remove cell debris for 24 hours, and pressure-selected with 1 μM puromycin for 1 week, and single clones were selected and transferred to 96-well plates for further culture for 2 weeks. image 3 As shown, the specific Myc monoclonal antibody was used to identify the Myc-biotididase-KRAS target band (55 kDa in size) in Western blotting, and cell clones highly expressing the fusion prote...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com