Primer probe for identifying novel coronavirus and application of primer probe in triple fluorescent RPA
A coronavirus and probe technology, applied in the field of applied biology, can solve the problem of not particularly good timeliness, and achieve the effect of low detection sensitivity and wide application prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Embodiment 1, the acquisition of primer, probe sequence and the construction of positive plasmid
[0047] According to the full-length nucleic acid sequence of the new coronavirus (SARS-CoV2), compare and analyze it, and compare it with the full-length sequence of SARS virus and SARS-like virus to find out the SARS-CoV2 virus-specific sequence S gene and virus conserved sequence N gene, SARS-like virus conservative sequence E gene, Orf1Ab gene, M gene, then design primers and probes according to the above sequence; the primers and probes involved in this application are shown in the following table:
[0048]
[0049]
[0050] This application completes the sensitivity test of the fluorescent RPA method by constructing a standard plasmid. Based on the pUC57 plasmid, the target gene was synthesized and the recombinant plasmid was constructed. Transform Escherichia coli (Escherichia coli) competent cells by heat shock method, smear a plate to select monoclonal colon...
Embodiment 2
[0051] Embodiment 2, the processing of actual detection sample and virus RNA genome extraction
[0052] Test samples include but are not limited to human serum, plasma, whole blood, feces, throat swabs, and sputum. Sample processing: Dissolve 1g of feces or a patient's throat swab in 1ml of normal saline, shake and mix well, centrifuge at 5000r / min for 10min, and take the supernatant to extract viral RNA. Add an equal amount of 10g / L acetylcysteine to the sputum, shake it at room temperature for 1 hour, and extract viral RNA after fully liquefied. Anticoagulants (heparin, etc.) should be added to plasma and whole blood.
[0053] The guanidinium isothiocyanate-glass powder method extracts viral RNA: the specific operation steps are as follows: (1) add guanidine isothiocyanate-glass powder lysate (4.7mol / L guanidine isothiocyanate, 20mmol / LEDTA, 100mmol / L Tris-HCl pH6.4, 1% Triton X-100 and glass powder 10g) 0.9ml, mix well, act at room temperature for 10min, centrifuge at ...
Embodiment 3
[0055] Embodiment 3, implementing triple fluorescent RPA
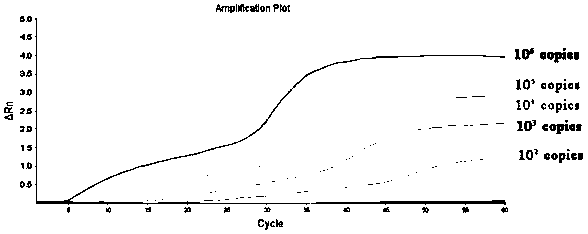
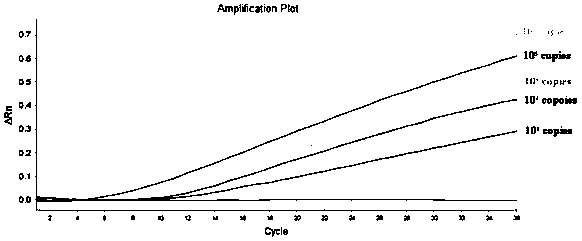
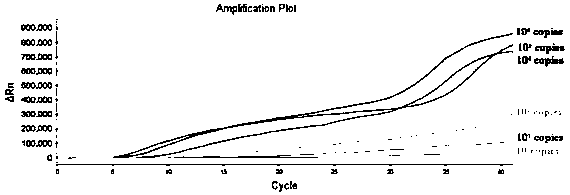
[0056] The triple fluorescent RPA reaction includes two steps: system preparation, amplification and signal reading. Use the 50 μL system recommended by the RNA Constant Temperature Rapid Amplification Kit (fluorescent type), use the prepared RNA or (DNA plasmid) as a template, and use the primers and probe sequences shown in the abstract to perform a triple fluorescent RPA reaction. Fluorescent RPA reaction system (50 μL): Buffer A29.4 μL, upstream primers NF, EF, SF each 0.8 μL (10 μmol / L), downstream primers NR, ER, SR each 0.8 μL (10 μmol / L), probes NP, EP , SP 0.3 μL each (10 μmol / L), nucleic acid template 2 μL, RNase-free Water 10.2 μL, ROX dye 0.2 μL, Buffer B 2.5 μL. The reaction conditions were set as follows: 42° C., 20 min, and the fluorescence signal was collected every 30 s, for a total of 40 cycles. The instrument automatically collects fluorescence signals, and can observe fluorescence amplification si...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com