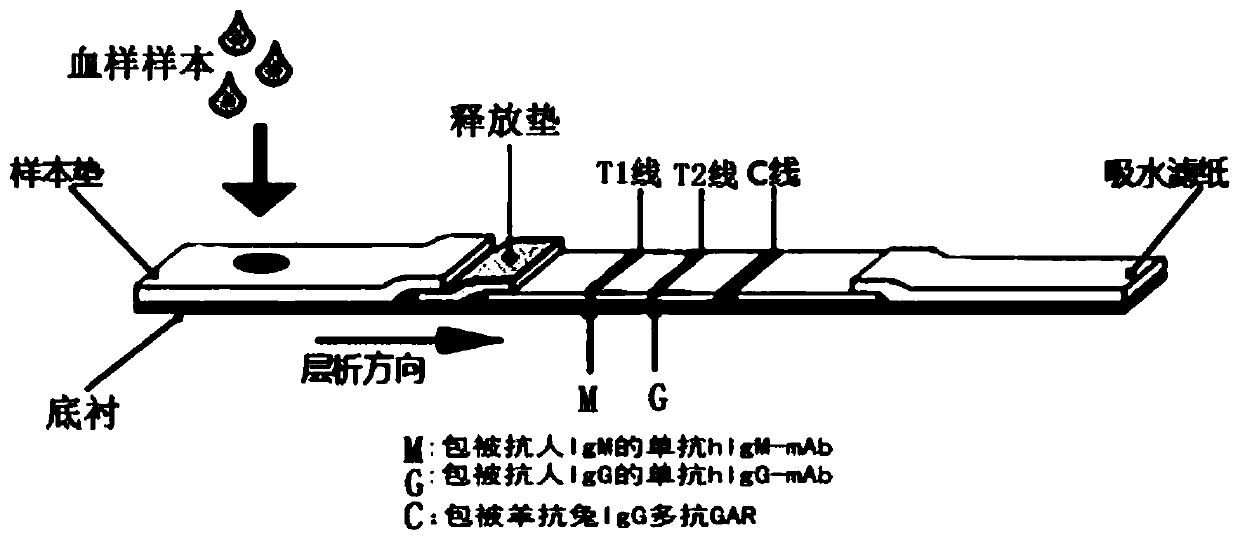
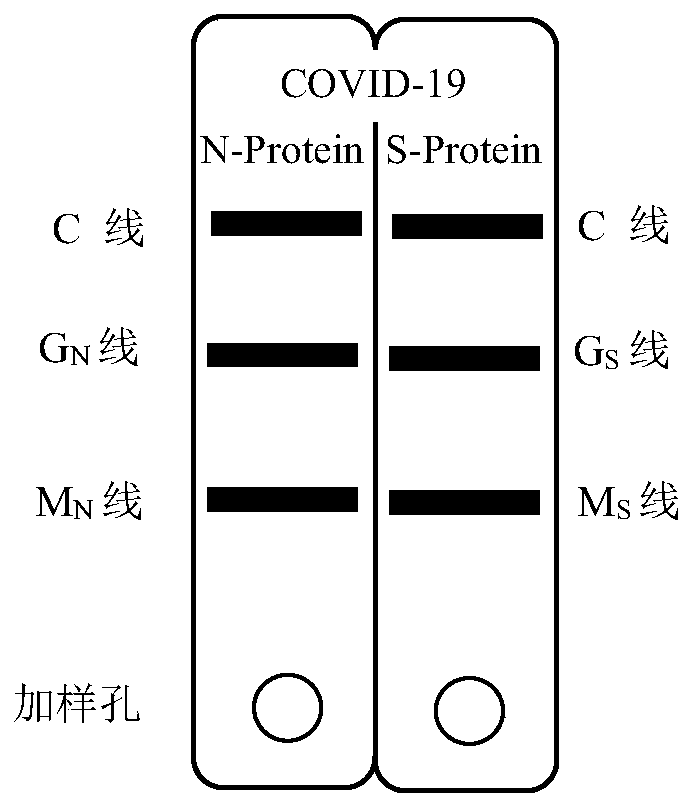
Novel coronavirus pneumonia (COVID-19) serological diagnosis kit
A technology of COVID-19 and diagnostic kits, which is applied in the field of serological diagnostic kits for novel coronavirus pneumonia, can solve the problems of very large differences in antibody levels and undetectable levels, so as to improve detection accuracy, improve detection sensitivity, and guarantee Biosecurity Effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
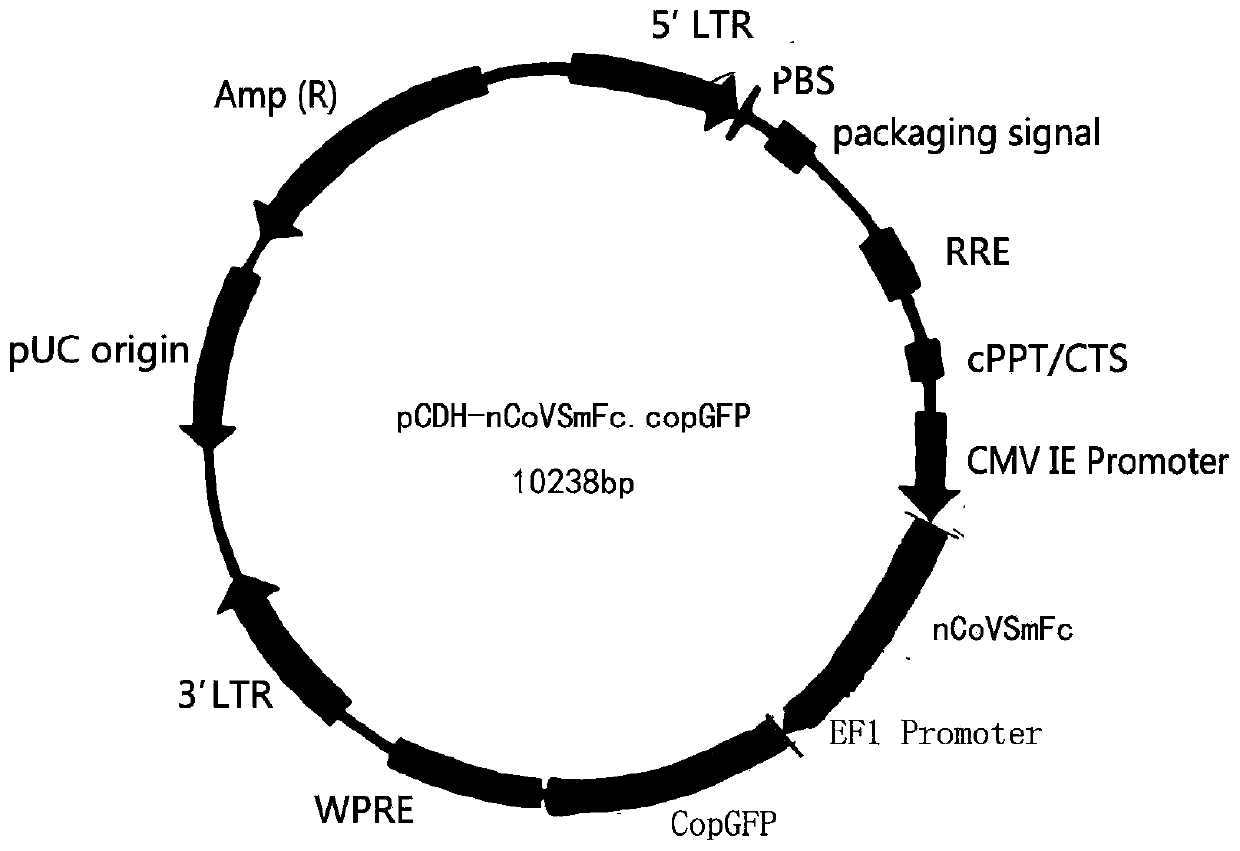
[0040] Example 1 pCDH-nCoVSmFc.copGFP plasmid construction
[0041] The C-terminal sequence of the outer part of the novel coronavirus spike protein is connected to the protein site-directed biotinylation sequence, a specific protease cleavage site tag and a purification tag to form an artificially designed sequence, wherein the outer part of the new coronavirus spike protein is NCBI Reference Sequence: 1-1213aa part of YP_009724390.1; the protein site-directed biotinylation sequence is biotin protein ligase BirA, its recognition site is GSGGSGGSAGGGLNDIFEAQKIEW, the specific protease is EK enzyme, and its cleavage site label is Flag Label DYKDDDDK, the purification label is mouse IgG Fc fragment (mFc).
[0042] Finally, the nucleic acid sequence of the gene-designed nCoVS-Flag-mFc fusion protein is obtained, and the host (human) codon-optimized sequence is shown in SEQ ID NO: 1, and EcoR I and Not I restriction endonucleases are respectively designed at both ends to express ...
Embodiment 2
[0049] Example 2 Establishment of nCoVSmFc.copGFP / 293 stably transfected cell line
[0050] The pCDH-nCoVSmFc.copGFP plasmid, pH1 plasmid, and pH2 plasmid were co-transfected into the lentiviral packaging line cell 293V to prepare nCoVSmFc.copGFP lentivirus, and transfected into HEK293 cells. The clones were picked under a fluorescent microscope to establish nCoVSmFc.copGFP / 293 stable transfected cell lines. Specific steps are as follows:
[0051] 1) The day before the experiment, five 15cm dishes were plated to ensure that the cells reached 70%-80% confluence before transfection.
[0052] 2) 1-2 hours before transfection, replace the medium in the dish with DMEM medium without serum and without double antibodies.
[0053] 3) Prepare a 15mL centrifuge tube, add 5mL 1×HBS, then add 100μg pCDH-nCoVSmFc.copGFP plasmid, 100μg PH1 / PH2 mixed plasmid (PH1:PH2=3:1, mass ratio), and mix gently .
[0054]4) Add 4mL PEI working solution (10μM), mix gently, and incubate at 37°C for 20...
Embodiment 3
[0065] Embodiment 3S-Biotin protein preparation
[0066] Use protein-free 293 cell culture medium to amplify and culture nCoVSmFc.copGFP / 293 stably transfected cells, collect supernatant culture fluid, and use protein A / G to purify nCoVSmFc fusion protein, use EK enzyme to cut nCoVS from the column, pass through biotin protein The ligase couples biotin to the (GSGGSGGSAGGGLNDIFEAQKIEW) site of nCoVS to obtain S-Biotin protein. Specific steps are as follows:
[0067] 1) Expand nCoVSmFc.copGFP / 293 stably transfected cells to a five-layer cell factory, and culture them in HektorHEK293 cell culture medium without proteins and peptides;
[0068] 2) Collect the culture medium, centrifuge at 1200g, 4°C for 20min, take the supernatant, and use saturated ammonium sulfate method to precipitate protein;
[0069] 3) Precipitated protein was reconstituted with PBS (pH7.4) of 1 / 10-1 / 100 volume of the original solution, desalted, concentrated, and replaced with binding / washing buffer (0.5M...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com