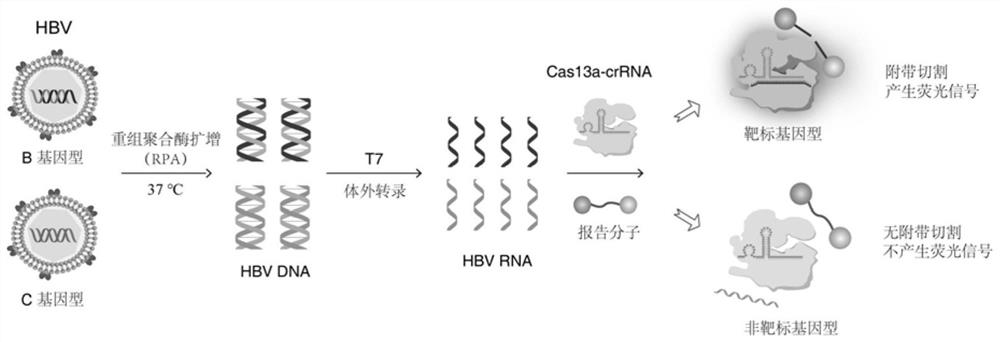
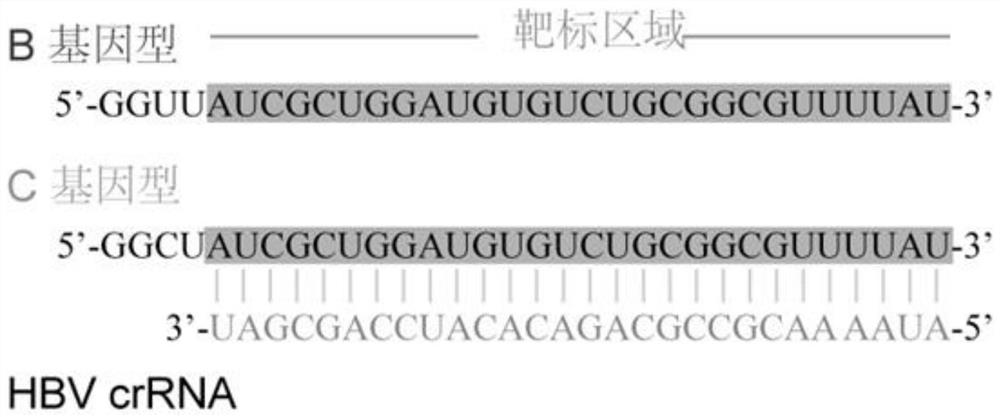
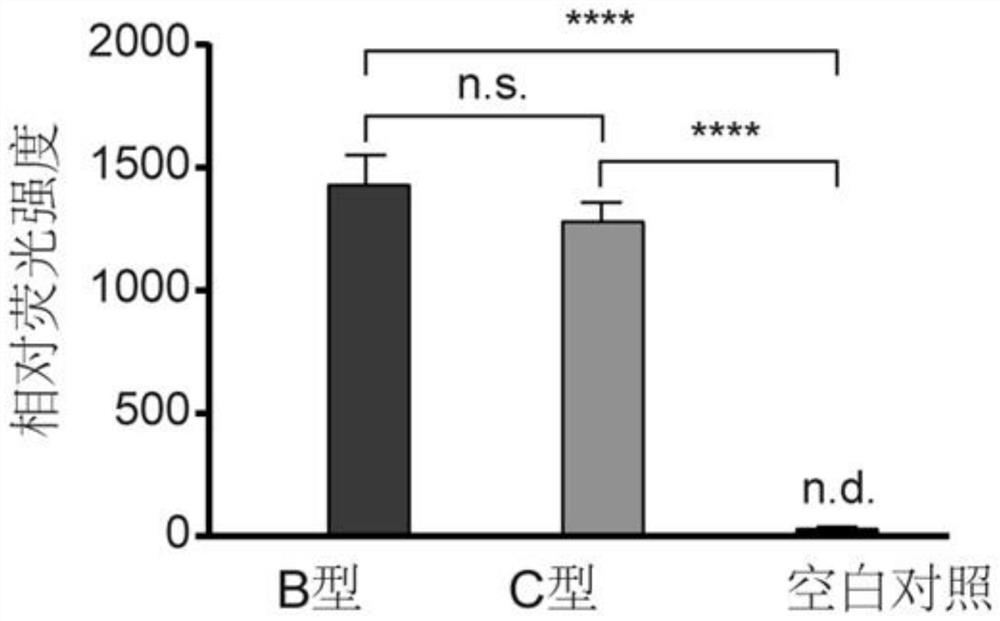
Hepatitis B virus genotyping detection method based on CRISPR/Cas13a system
A hepatitis B virus and genotyping technology, which is applied in the field of medical biology, can solve the problems of low detection sensitivity, difficulty, and poor stability, and achieve the effects of improving detection specificity, saving time and cost, and eliminating the need for operating procedures
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Design of RPA Primers for Genomic DNA of HBV Virus
[0052] 1. Use the NCBI Primer Blast online tool to design universal recombinant polymerase amplification (RPA) primers for HBV genomic DNA, which are effective for both HBV type B and C viruses, that is, only this pair of RPA primers is needed Amplification of HBV type B and C viruses.
[0053] 2. After screening, the final RPA primer pair used in the present invention is as follows: the sequence of the forward primer is shown in SEQ ID NO:1 and the sequence of the reverse primer is shown in SEQ ID NO:2.
Embodiment 2
[0055] Design crRNA sequences and synthesize them by in vitro transcription
[0056] 1. First synthesize a DNA sequence complementary to crRNA as a template for in vitro transcription. It should be noted that a complementary sequence of the T7 promoter should be extended at the 3' end of the DNA sequence, as shown in SEQ ID NO: 13, which is the subsequent T7 Prepare for in vitro transcription.
[0057] 2. Then anneal the DNA template and a short T7 promoter sequence, as shown in SEQ ID NO: 14, in 1x annealing buffer (100mM Tris-HCl (pH 7.5), 10mM EDTA, 1M NaCl), Incubate first at 95 °C for 10 min, then allow to cool slowly to room temperature.
[0058] 3. After the annealing reaction, use the HiScribe T7 Rapid High-yield RNA Synthesis Kit to incubate overnight at 37°C for in vitro transcription of crRNA. The transcription reaction contained NTP buffer mixture (the final concentration of each NTP was 10 mM), T7 RNA polymerase mixture, template DNA (2 μg), mouse RNase inhibito...
Embodiment 3
[0061] Synthesis and purification of HBV RNA
[0062] 1. First, use the designed RPA primer pair to amplify the plasmid containing HBV DNA through the RPA kit. The amplified product was then incubated overnight at 37°C for in vitro transcription using the HiScribe T7 Rapid High Yield RNA Synthesis Kit.
[0063] 2. After the in vitro transcription reaction is completed, add an appropriate amount of RNase-free DNase I (4 units in total) to the system and incubate at 37°C for 15 minutes to digest the DNA template.
[0064] 3. The obtained HBV in vitro transcription product was purified by MEGAclear transcription purification kit, and then the concentration was measured by Nanodrop, and verified by denaturing urea polyacrylamide gel electrophoresis (urea / PAGE).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com