Larimichthys crocea genome breeding chip and application
A large yellow croaker and genome technology, applied in the fields of molecular biology and genome breeding, bioinformatics, and genomics, can solve the problems that restrict the healthy and sustainable development of the large yellow croaker farming industry, the decline of genetic diversity of the cultured population, and the lack of germplasm resources and other issues, to achieve the effects of reduced sequencing costs, improved sequencing throughput, and simple data analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1 Preparation method of "Ningxin No. 1" large yellow croaker genome breeding chip
[0043] 1. Using Illumina sequencing technology, the whole genome of 82 large yellow croakers from 5 groups was resequenced. The total sequencing volume was 650.5G, and the average sequencing depth of each sample was 11.33X (Table 1). After aligning the sequencing data to the high-quality large yellow croaker reference genome sequence ([19] Chen B H, Zhou Z X, Ke Q Z, Wu Y D, Bai H Q, Pu F, XuP. The sequencing and de novo assembly of the Larimichthys crocea genome using PacBio and Hi-C technologies [J]. Scientific Data, 2019, 6), follow the steps below to identify and screen SNPs.
[0044] Table 1 The amount of sequencing and the number of SNPs in the population
[0045]
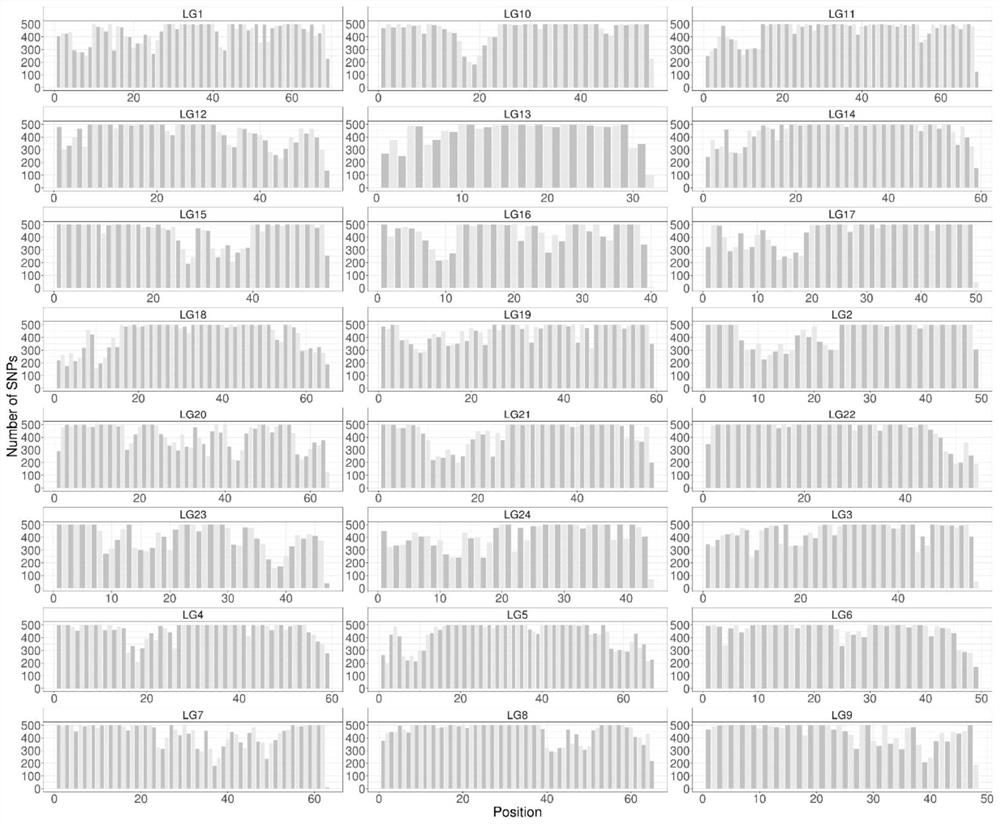
[0046] 2. Screen out 9,335,807 high-quality SNP sites from all large yellow croaker sequencing data. High-quality SNP sites are based on the following principles: the SNP sequence is located in a specific seq...
Embodiment 2
[0051] Example 2 Application of "Ningxin No. 1" large yellow croaker genome breeding chip in the detection of large yellow croaker DNA samples
[0052] (1) Genomic DNA extraction from large yellow croaker: Genomic DNA was extracted from large yellow croaker muscles, fin rays, blood and other tissues using the phenol-chloroform method or a DNA extraction kit (DNeasy 96 Blood and Tissue Kit, Qiagen, China) according to the detection requirements.
[0053] (2) DNA sample quality detection: detect with the agarose gel electrophoresis that mass fraction is 1-1.5% (W / W), judge electrophoresis result with gel imaging system (GelDocXRSystem, U.S. Bio-Rad company), guarantee genome DNA integrity is good, and the length of the genomic DNA fragment is greater than 10kb; measure the concentration of genomic DNA with a micro-volume ultraviolet spectrophotometer (Q5000, Quawell, USA) or a similar nucleic acid and protein analyzer, and adjust the DNA concentration to a working concentration o...
Embodiment 3
[0056] Example 3 Application of "Ningxin No. 1" large yellow croaker genome breeding chip in genetic background analysis of large yellow croaker breeding materials
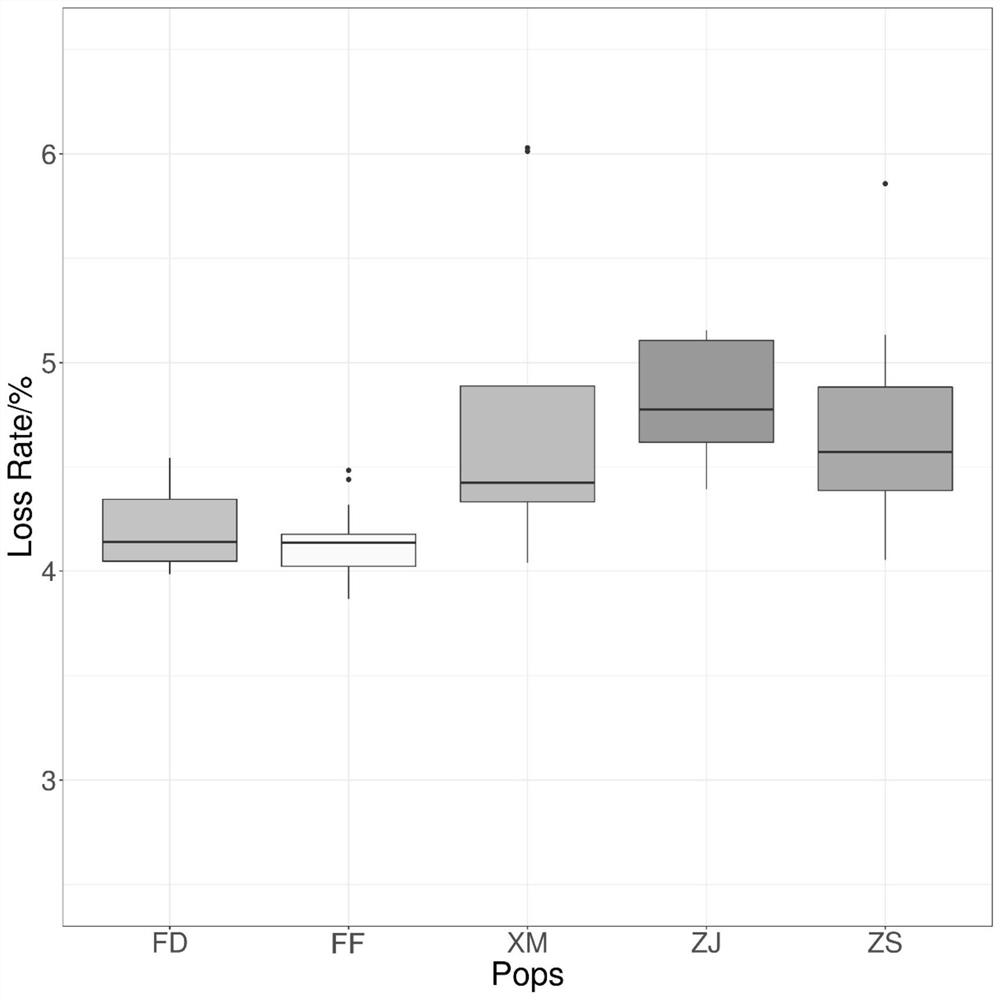
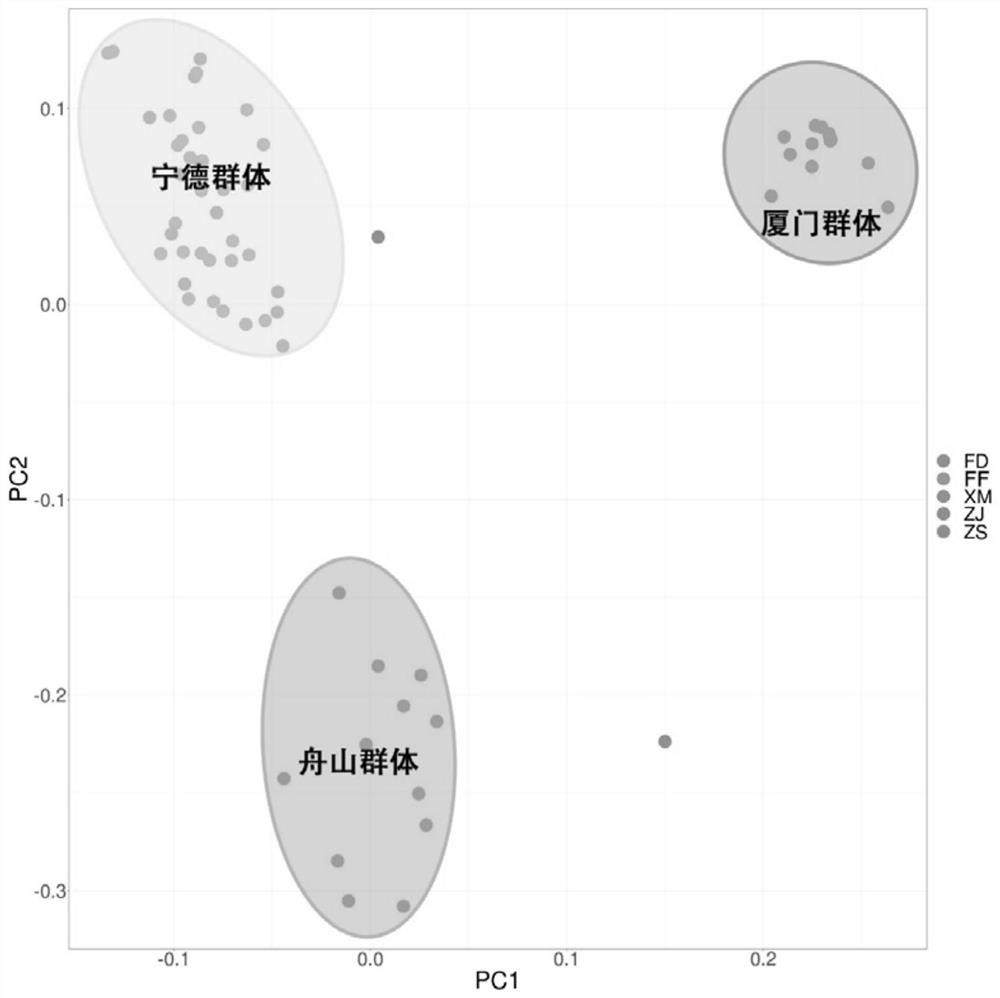
[0057] In order to verify the application of the "Ningxin No. 1" large yellow croaker genome breeding chip in the genetic background analysis of large yellow croaker breeding materials, the present invention was used to analyze samples from Zhoushan (12), Fuding (12), Xiamen (6), The samples of wild large yellow croaker from four locations in Zhanjiang (9) and 27 cultured large yellow croakers from Ningde City Fufa Aquatic Products Co., Ltd. were detected. The detection method refers to Example 2, and the results show that the typing success rate in all groups is average. Above 95% ( figure 2 ). After removing sites with a typing success rate of less than 90% and a minimum allele frequency of less than 2%, the genotype information of 514,726 high-quality SNP sites was obtained. Based on this, the principal comp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com