Chromosome copy number variation detection device based on low-depth high-throughput genome sequencing
A chromosome copy number and genome sequencing technology, applied in the field of bioinformatics, can solve problems such as large impact on data quality, sequence-specific deviation, library preparation deviation, etc., and achieve the effect of obvious CNV breakpoints, stable data, and accurate results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] This embodiment provides a method for detecting chromosomal variation using the device for detecting chromosomal copy number variation based on low-depth high-throughput genome sequencing of the present invention.
[0073] The device includes: a detection module, a data quality control module, a data preprocessing module, a data correction and processing module and a judging module. Specific steps are as follows:
[0074] 1. Negative reference set construction
[0075] (1) Sample selection
[0076] A total of 300 peripheral blood samples from healthy people with no chromosomal abnormality in karyotype analysis were selected, and there was no significant difference in the proportion of males and females. DNA was extracted, genome sequencing was carried out according to the high-throughput method, and fastq data with a read length of 50 bp was obtained by single-end sequencing. The sequencing platform used was the MGISEQ-2000 gene sequencer manufactured by MGI.
[0077...
Embodiment 2
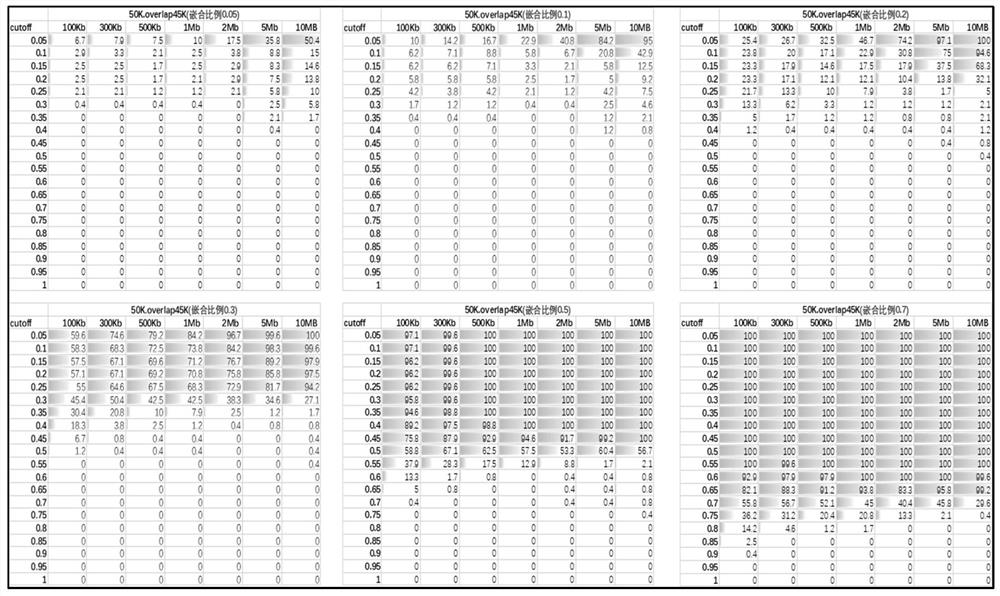
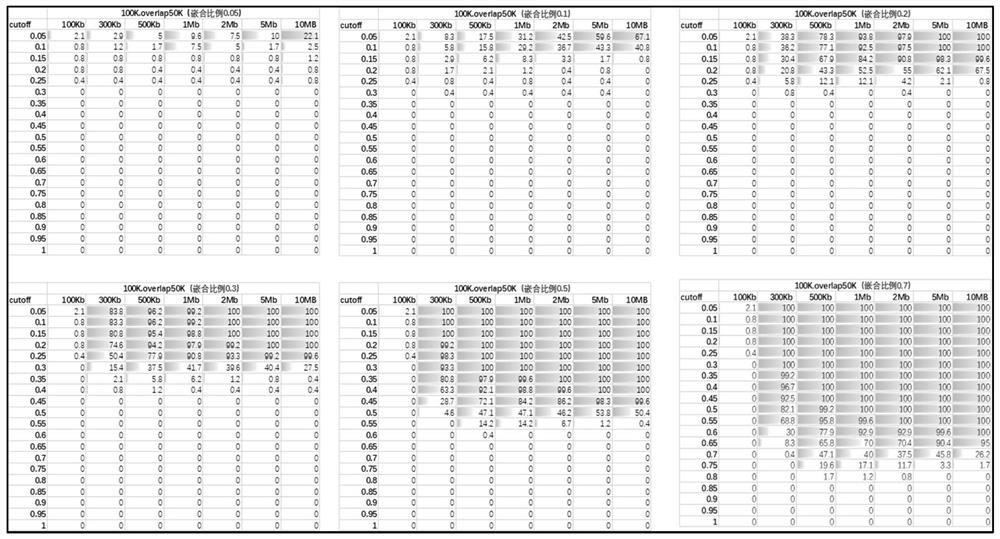
[0134] In this embodiment, the method shown in Embodiment 1 is used to test the experimental mixed sample data.
[0135] 13 positive samples with chip results and different sizes of abnormal fragments were mixed according to a certain chimerism ratio for library construction, chimerism gradient: 10%, 20%, 40%, 60%, 80%, 100%; positive region information As shown in table 2.
[0136] Table 2
[0137]
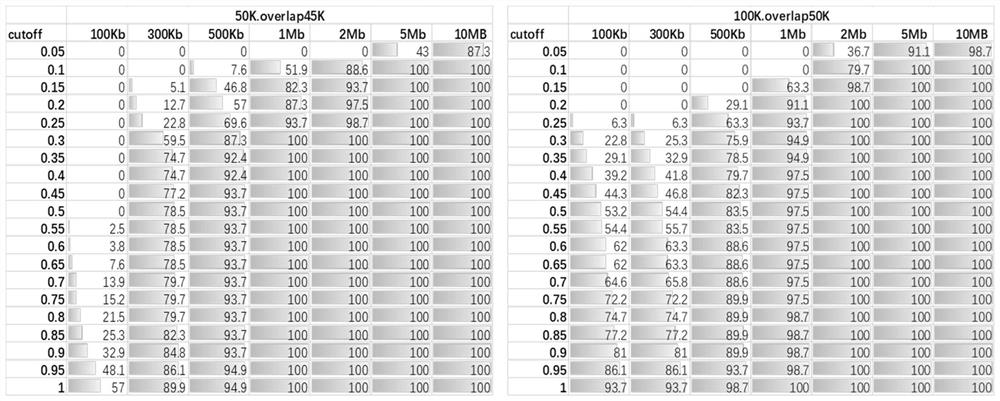
[0138] Specifically according to the method of the present invention, when the window is divided into 50kb, the R of the candidate CNV region of the above-mentioned positive sample obtained sample 、mean R reference 、sd R reference , Z value see Table 3. When the window is divided into 100kb, the R of the candidate CNV regions of the above positive samples obtained sample 、mean R reference 、sd R reference , Z value see Table 4. According to the judgment method of the present invention, the judgment results obtained under the two window widths are integrated, and the fin...
Embodiment 3
[0149] In this embodiment, the method shown in Embodiment 1 is used to test the clinical sample data.
[0150] Select 20 clinically positive samples (confirmed by CytoScan 750K SNP-Array chip test), including 8 males and 12 females. The sample types include: embryonic tissue, abortion tissue, villi, and induced fetal tissue. The specific sample types and chip results are shown in Table 5. The above 20 samples were detected by the method of Example 1. The two judgment results obtained after dividing according to the size of the two windows are combined, and the final judgment results obtained are all positive. The test results are shown in Table 6.
[0151] table 5
[0152]
[0153] Table 6
[0154]
[0155] According to the above results, it can be seen that the method of the present invention is used to detect all the small fragment CNVs above 100kb, all the CNVs and aneuploids with a low proportion of mosaicism, and all the regions with multiple abnormal CNVs. The ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com