Amino acid sequence determination method based on quasi-isobaric double labeling at two ends of polypeptide
An amino acid, double-labeling technology, applied in measurement devices, biological testing, material analysis by electromagnetic means, etc., can solve the problems of inability to simplify the spectrum, low B ion response, complex spectrum, etc., to achieve reaction efficiency and selectivity High, improved accuracy, and improved specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] 1. Isotope-labeled peptide ends based on free solution
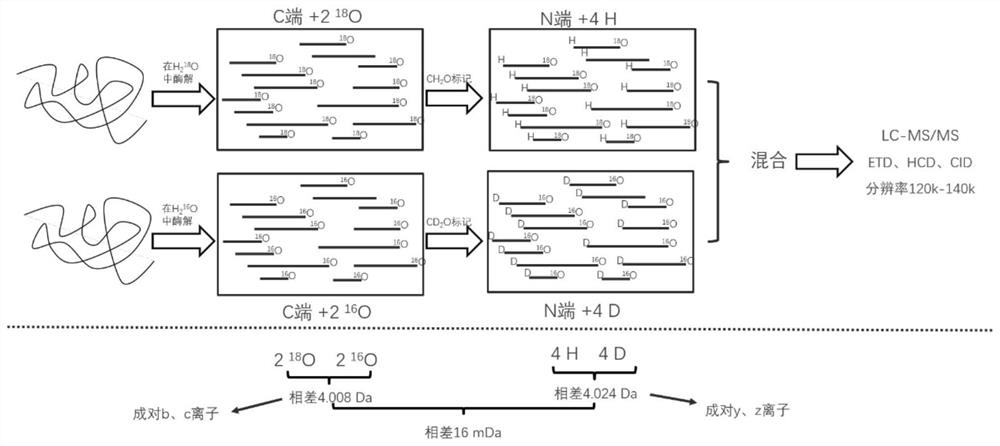
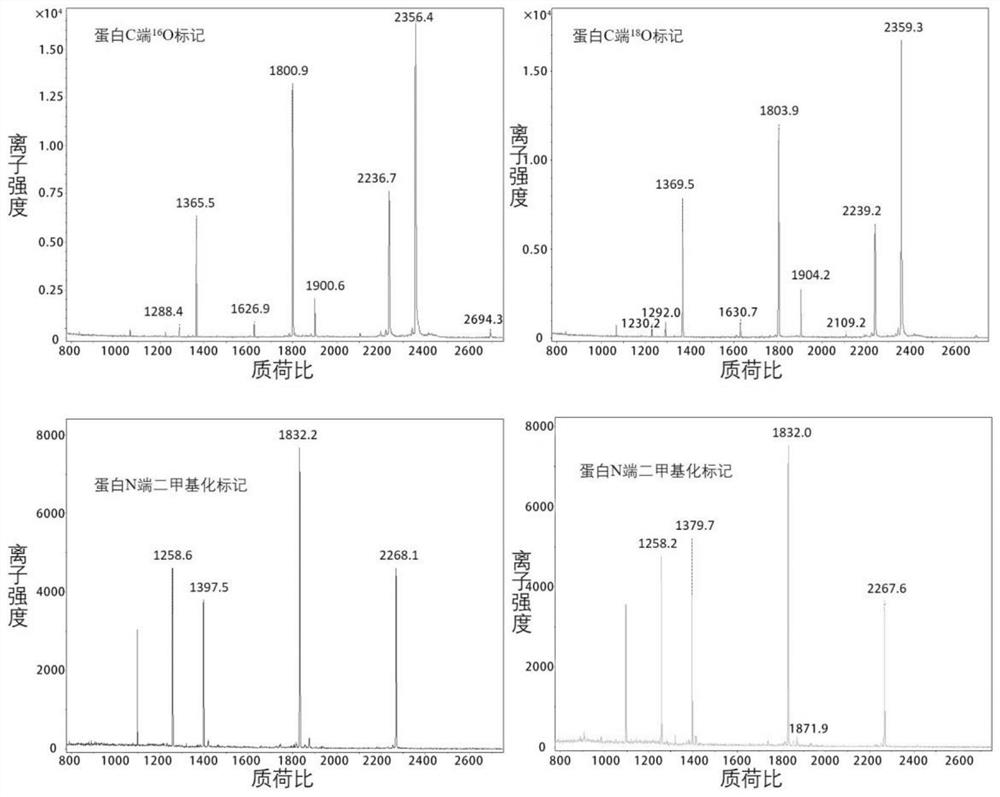
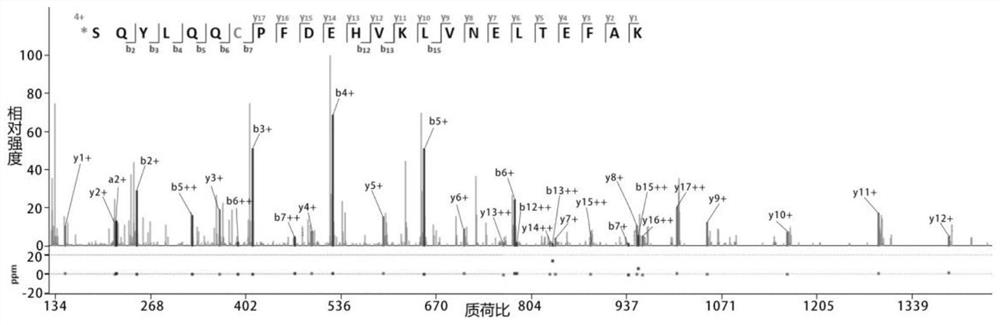
[0025] Such as figure 1 As shown, mark according to the following process.
[0026] (1) Denaturation, reduction, alkylation and enzymatic hydrolysis of protein samples: 1 mg of bovine serum albumin (BSA) was dissolved in 1000 μl of 50 mM ammonium bicarbonate solution, with a final concentration of 1 mg / ml. Heat at 95°C for 15 minutes to denature BSA, add 8 μl of 1M DTT, mix well, heat at 56°C for 1 hour; then add 20 μl of 1M IAA, and store in the dark for 30 minutes. Add 20 μg of trypsin with a concentration of 1 mg / ml into the centrifuge tube, mix well, heat in a water bath at 37° C. for 12 hours, and add 2 μl of formic acid to stop the enzymatic hydrolysis. Use a C18 reverse-phase chromatographic column to desalt and freeze-dry to obtain trypsin-digested polypeptide powder.
[0027] (2) Labeling at both ends of the peptide in the free solution: take 100 μg of the above peptide powder, place it in a 500 μl cen...
Embodiment 2
[0031] Based on the immobilized enzyme reactor and chemically labeled protein enzymatic digestion polypeptide both ends of the label.
[0032] In this example, different methods of labeling the C-terminus of the polypeptide are adopted, and the methods for labeling, separating and identifying the N-terminus of the polypeptide are the same as those in Example 1 above.
[0033] 1) Denaturation, reduction, alkylation, enzymatic hydrolysis and C-terminal labeling of protein samples: 1 mg of protein was dissolved in 1000 μl of 50 mM ammonium bicarbonate solution, with a final concentration of 1 mg / ml. Heat at 95°C for 15 minutes to denature BSA, add 8 μl of 1M DTT, mix well, heat at 56°C for 1 hour; then add 20 μl of 1M IAA, and store in the dark for 30 minutes. The denatured and reductively alkylated protein was divided into two parts and freeze-dried. Use H respectively 2 16 O and H 2 18 O The concentration is 50 mM ammonium acetate aqueous solution to dissolve the powder, t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com