Indirect ELISA kit for detecting feline coronavirus antibody
A feline coronavirus and kit technology, applied in viral peptides, biological testing, introduction of foreign genetic material using vectors, etc., can solve the problems of high S1 functional domain variability and unfavorable target protein regions, and achieve strong specificity and sensitivity. High and stable effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Cloning sequencing analysis, expression and purification of embodiment 1 feline coronavirus S recombinant protein
[0033] (1) Primer design and synthesis
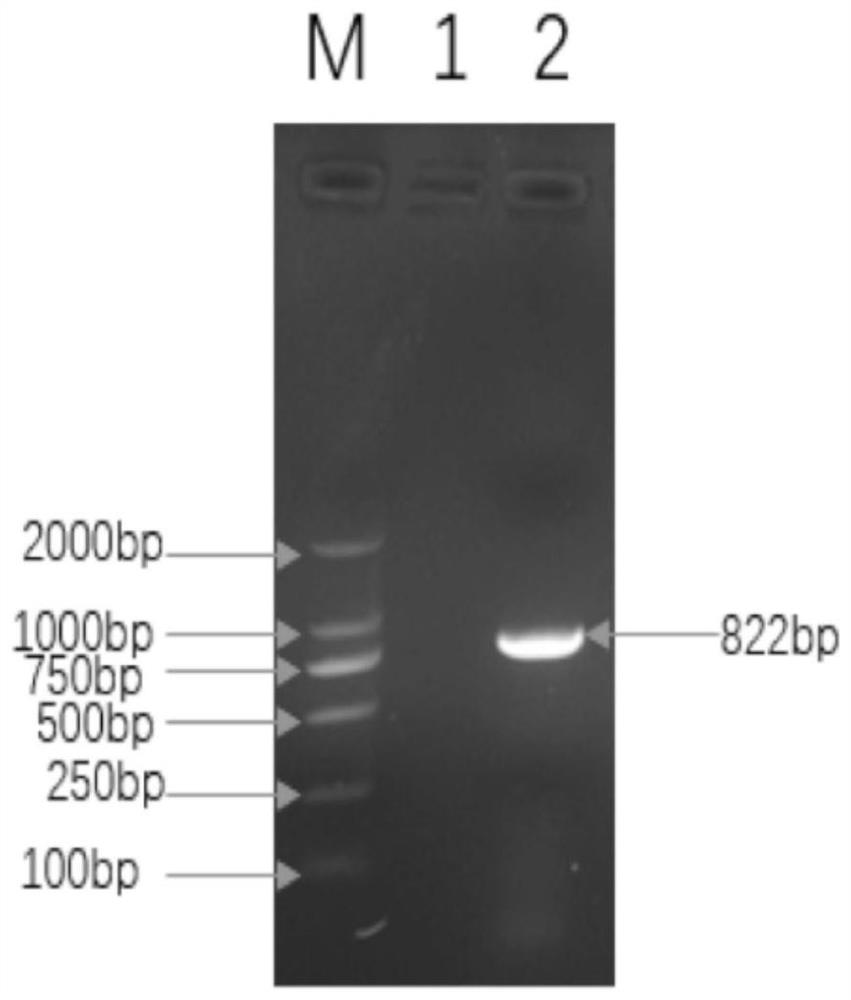
[0034] Referring to the FCoV S protein gene sequence published in GenBank, a pair of specific primers were designed and synthesized: the upstream primer sequence is shown in SEQ ID NO:2; the downstream primer sequence is shown in SEQ ID NO:3. The double enzyme cutting sites in the downstream primers were designed as NdeI and XhoI respectively. Primers were synthesized, purified and sequenced by Shanghai Sangon Bioengineering Co., Ltd.
[0035] (2) PCR using the above primers
[0036] Feline coronavirus S protein gene was used as template, the amplification system was 25 μL, including 12.5 μL of 2×TransStart FastPfuPCR SuperMix, 1 μL of upstream and downstream primers, 1 μL of plasmid template, supplemented with sterilized ddH 2 O to a final volume of 25 μL. Reaction conditions: Pre-denaturation at 94°C for 2 min...
Embodiment 2
[0040] Reactogenicity and immunogenicity identification of embodiment 2S recombinant protein
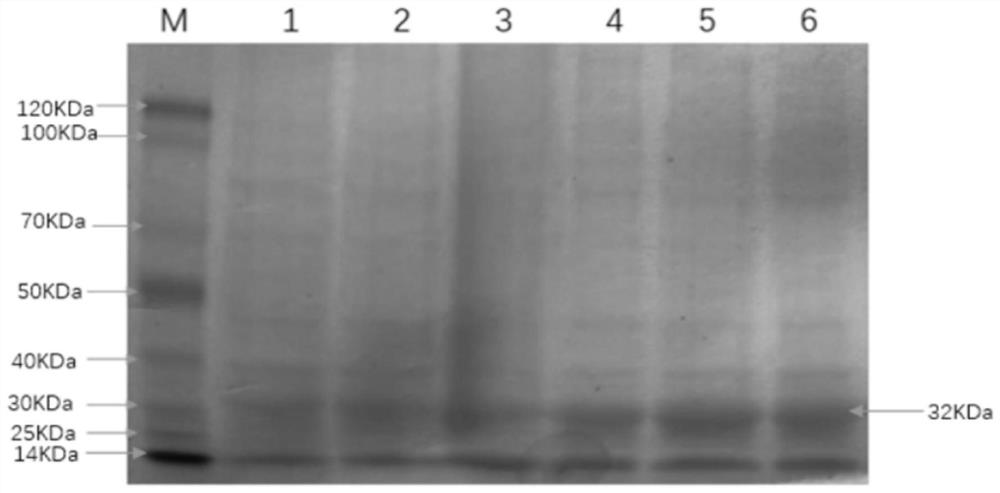
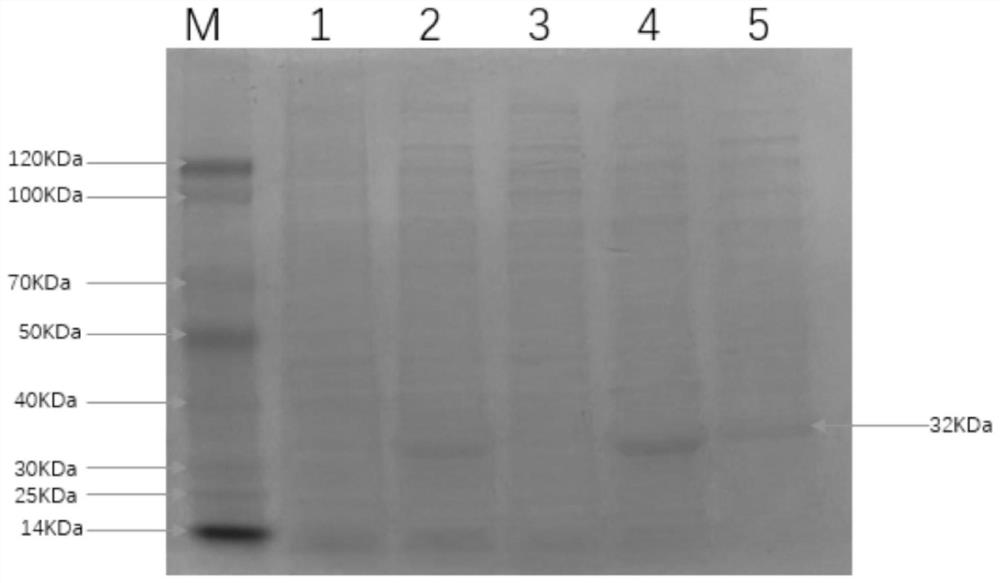
[0041] Firstly, the above-mentioned bacterial liquid was ultrasonicated and then centrifuged to obtain inclusion body precipitation, and the inclusion body was denatured. The denatured supernatant was passed through His-Trap column affinity chromatography, and the liquid after passing through the column was collected, and then subjected to gradient dialysis with refolding buffer, and finally Collect the supernatant. After performing SDS-PAGE electrophoresis on the purified recombinant fusion protein, the electrophoresis results are as follows: image 3 As shown, electrotransfer to nitrocellulose membrane for Western blot identification, such as Figure 4 As shown, the results show that the feline coronavirus S recombinant protein of the present disclosure can react with cat serum and has biological reactogenicity. The purified and identified recombinant protein was dialyzed with PB...
Embodiment 3
[0042] Embodiment 3 is used to detect the development of the indirect ELISA kit of cat coronavirus antibody
[0043] 1. Determination of the optimal coating concentration of the antigen and the optimal concentration of the secondary antibody
[0044] (1) Dissolve the purified protein in the coating solution at an appropriate concentration, and start from left to right for doubling dilution (8μg / ml, 4μg / ml, 2μg / ml, 1μg / ml, 0.5μg / ml ml, 0.25 μg / ml). The antigen was added to the ELISA plate at 50 μl per well for overnight coating at 4°C.
[0045] (2) Take out the coated ELISA plate, pour off the liquid in the plate, and empty it, wash it with PBST (200μl) for 3 times, each time for 5min, use 5% skimmed milk to press 100μl / well, at 37℃ Closed for 2 hours.
[0046] (3) Take out the ELISA plate that has been blocked, pour off the liquid in the plate, and empty it, wash it with PBST (200μl) for 3 times, each time for 5min, and use 50μl / well of positive serum (primary antibody) at ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com