Method for preparing small-amount sample high-throughput chromatin co-immunoprecipitation sequencing sample
A co-immunoprecipitation and sample preparation technology, which is applied in the field of sample preparation for high-throughput chromatin immunoprecipitation sequencing of a small number of samples, can solve the problems of a large number of initial samples and low throughput, and achieve the effect of low price and simple processing technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0086] Example 1. Sample preparation process based on the novel superhydrophobic microwell array chip high-throughput chromatin immunoprecipitation-sequencing (ChIP-seq) technology
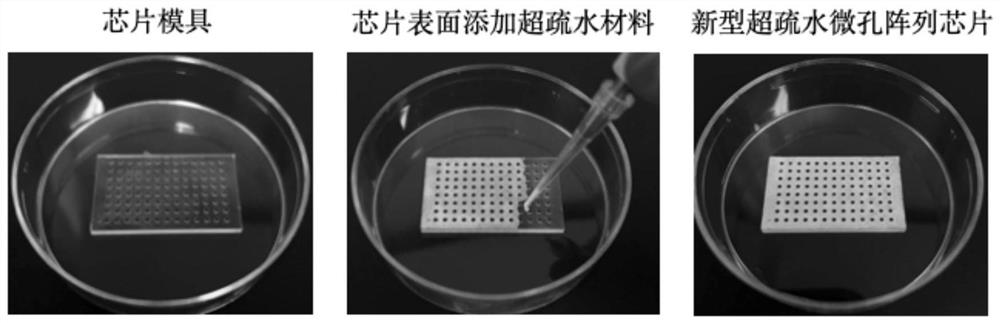
[0087] First of all, the processing of the new super-hydrophobic microhole array chip is carried out, as follows:
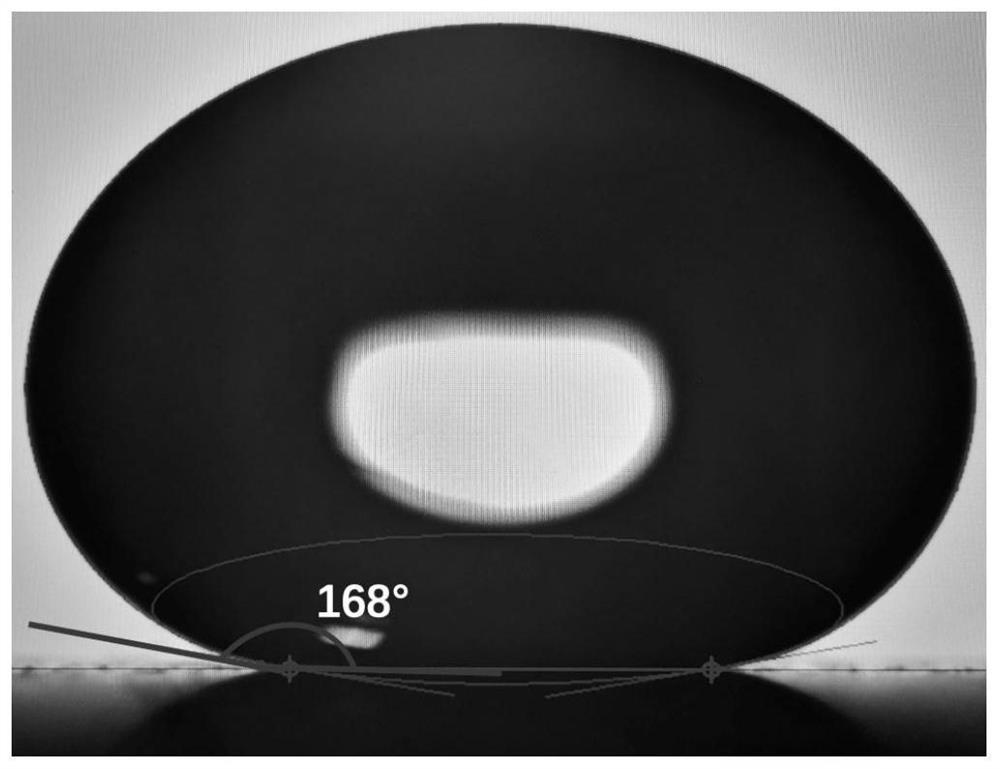
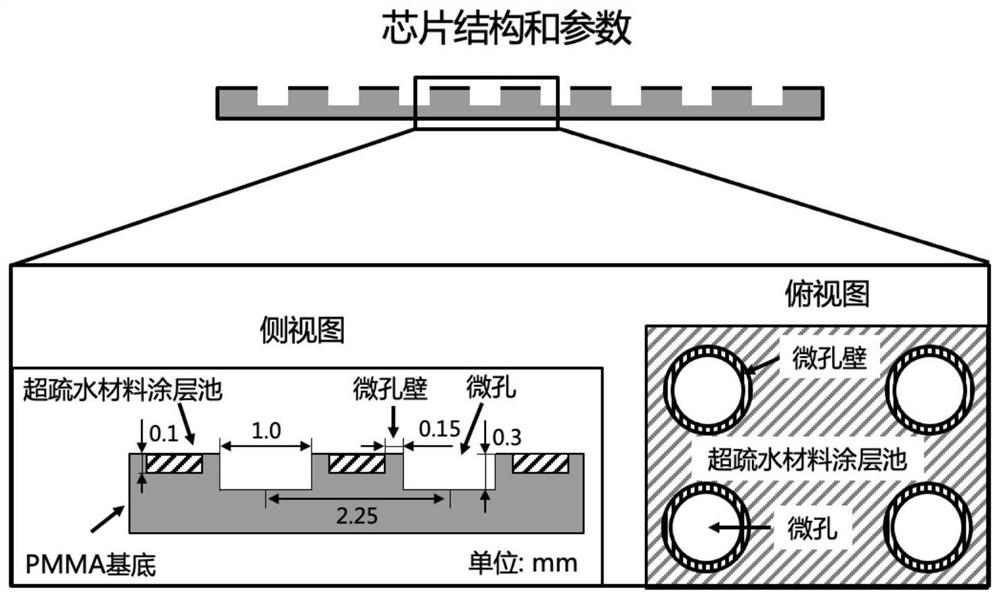
[0088] 1) PMMA mold design. The mold material is PMMA, and there are 108 microhole arrays (12×9) designed on the mold. The microholes are cylindrical, and the microholes are evenly distributed in the microhole array. is 2.25mm. A superhydrophobic material coating pool is designed between the micropore arrays, the micropores and the micropore wall periphery are all the coating pools, and the depth of the coating pool is 0.1mm. The micropore wall thickness is 0.15mm.
[0089] 2) Synthesis of novel superhydrophobic coatings. 2g 1H, 1H, 2H, 2H-perfluorooctyltriethoxysilane (Sigma, 718467) was mixed with 198g absolute ethanol for 2 hours under mechanical stirring, and 10g of titanium ...
Embodiment 2
[0103] The specific application example of embodiment 2, embodiment 1 method
[0104] 1. Using the method in Example 1, the H3K4me3 ChIP-seq of 100 samples and 1000 K562 cells in each sample is completed based on the chip. Since each microwell is independently coded, it can be split after sequencing Samples with different encodings were obtained, and compared with the K562 cell H3K4me3 ChIP-seq results in the Encode database, there was good consistency. For specific results, see Figure 6 .
[0105] 2. Adopt the method in Example 1 to implement four different histone modifications based on the chip (H3K4me1 (Abcam, ab8895); H3K4me3 (Millipore, 07-473); H3K27ac (Abcam, ab4729) and H3K27me3 (Millipore, 07-449 )) for ChIP-seq sample preparation with 24 microwell replicates for each histone modification, with an initial volume of approximately 1000 K562 cells per sample. For specific results, see Figure 7 . in, Figure 7 Middle a is a schematic diagram of the seeding distri...
PUM
| Property | Measurement | Unit |
|---|---|---|
| depth | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| depth | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com