Keratinase mutant with improved thermal stability and application
A kind of keratinase mutation, keratinase technology, applied in the application, hydrolase, microorganism-based method and other directions, can solve the problem of time-consuming and laborious, the protein structure-activity relationship is not completely resolved and other problems, and achieve the effect of good application value and prospect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment 1: Construction of keratinase homology model
[0035] According to the amino acid sequence of the keratinase gene kerBp, the homology comparison was carried out in the NCBI database, and three crystal structures with high homology (greater than 60%) were selected as templates, and the 3D three-dimensional structure of the keratinase was simulated by EasyModeller software, and SAVES was used. The Verify 3D function of the online analysis server (http: / / servicesn.mbi.ucla.edu) evaluates the quality of the built model. The relevant analysis operation of the three-dimensional structure was performed by PyMOL and DiscoveryStudio software.
Embodiment 2
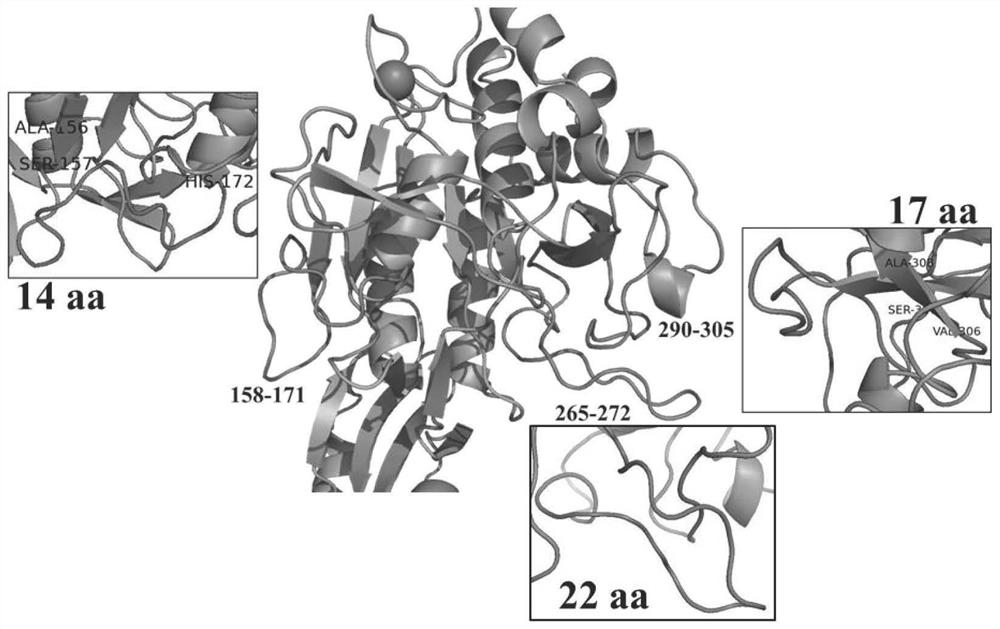
[0036] Example 2: Calculation and Analysis of Highly Flexible Loop Area of Keratinase
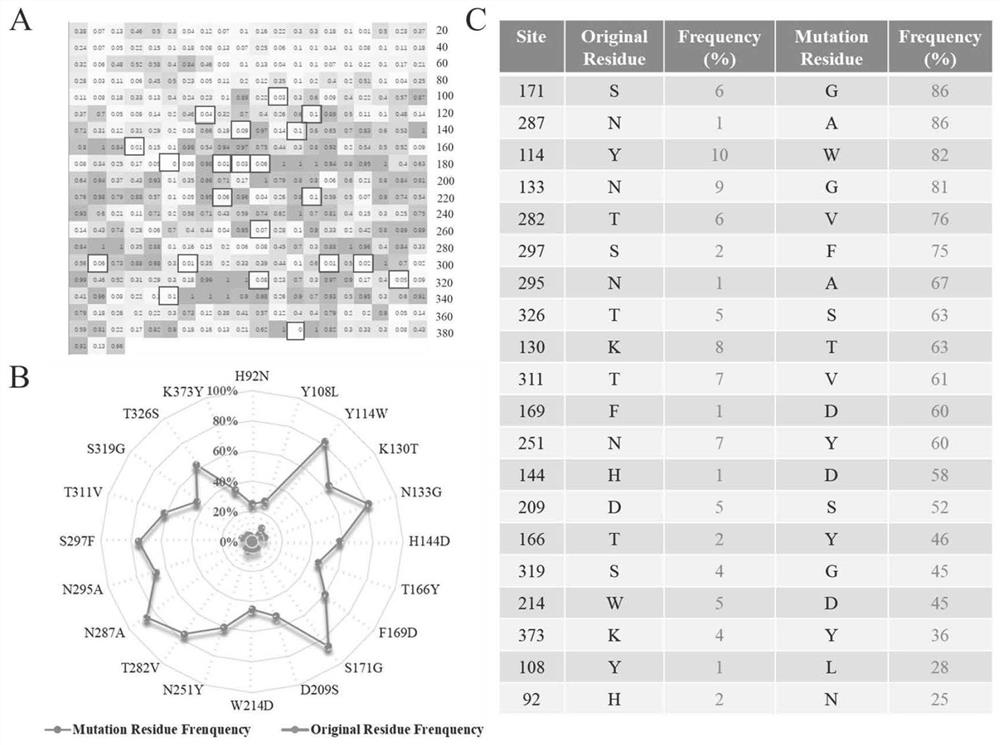
[0037] The sequence (SEQ ID NO.2) and structure of keratinase were analyzed by using FoldUnfold and IUPred protein disorder region structure prediction tools. Combined with the homologous sequence comparison analysis and the visual analysis of the structure, three highly flexible Loop regions were determined, namely Loop158-171, Loop265-272 and Loop290-305 ( figure 1 ). Further, the B-Factor and RMSF of the amino acids in the above three Loop regions were predicted and calculated by the prediction software PredyFlexy, and 22 amino acid residues with higher B-Factor and RMSF were screened out as the objects of site-directed mutation. Based on the 22 mutation sites on the 3 highly flexible loops screened above, this experiment judged whether the mutant type was stable by calculating the difference between the Gibbs free energy change (ΔΔG) between the wild type and the mutant type, and fur...
Embodiment 3
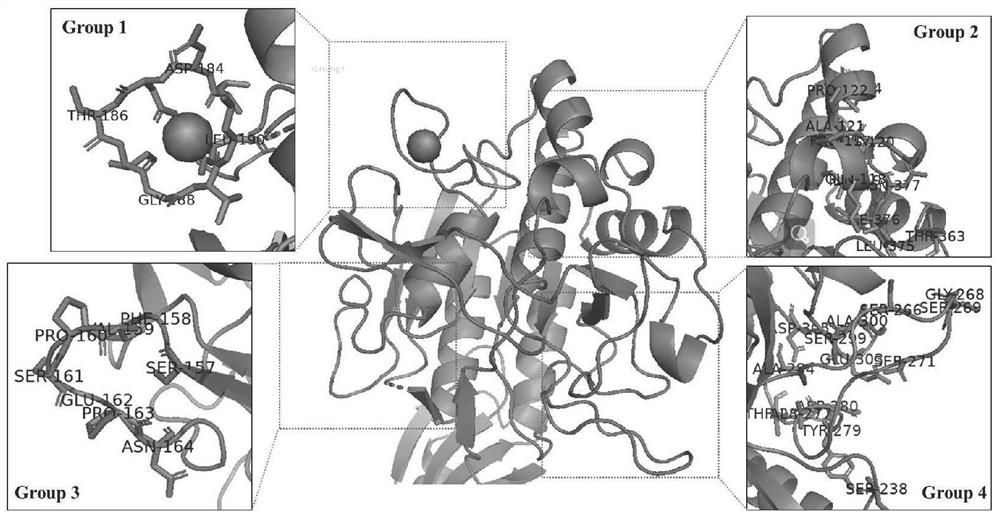
[0038] Example 3: Prediction and analysis of the calcium ion binding region of keratinase
[0039] The amino acid sequence of keratinase was analyzed by the online prediction tool IonCom, and 37 potential amino acid residues related to calcium ion binding were predicted, and these amino acids were further positioned in the homology model structure of keratinase and divided into three parts from the spatial position 4 groups ( figure 2), by observing the characteristics of the three-dimensional structure of the above four groups, a conserved calcium ion binding site CaI composed of amino acids D149, L183, D184, N185, T186, I187, G188, V189 and L190 was determined. According to the analysis of the CaI region of the conserved calcium ion binding site of keratinase, D149, L183, I187, V189 and N185 are the conserved amino acids of the calcium ion binding site of keratinase, in which the calcium ion directly interacts with residues D149, L183, I187 and V189 Coordination, while oth...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com