Saccharomyces strain for degrading organic acids and application of saccharomyces strain
A technology of yeast strains and organic acids, applied in the direction of yeast-containing food ingredients, fungi, and methods based on microorganisms, which can solve problems such as poor taste
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 Screening and Identification of a Yeast Strain Degrading Organic Acids
[0025] The gene sequence of the dominant strain that efficiently degrades organic acids in berries is shown in the sequence table SEQ ID NO.1; the dominant strain is similar to Zygosaccharomyces ( Zygosaccharomyces bisporus ) has the closest genetic distance and the best homology, combined with the strain's morphological characteristics and physiological and biochemical tests, it can be judged that the strain is Zygomyces dispora ( Zygosaccharomyces bisporus ); the dominant strain was deposited in the China Center for Type Culture Collection in Wuhan, Hubei Province on December 16, 2020, with the preservation number CCTCC M 2020912. The screening and identification methods of strains are as follows:
[0026] 1. Separation and purification of bacteria
[0027] Take 4.9g of yeast extract powder peptone glucose agar medium and dissolve it in 100mL of distilled water, put it in an autocl...
Embodiment 2
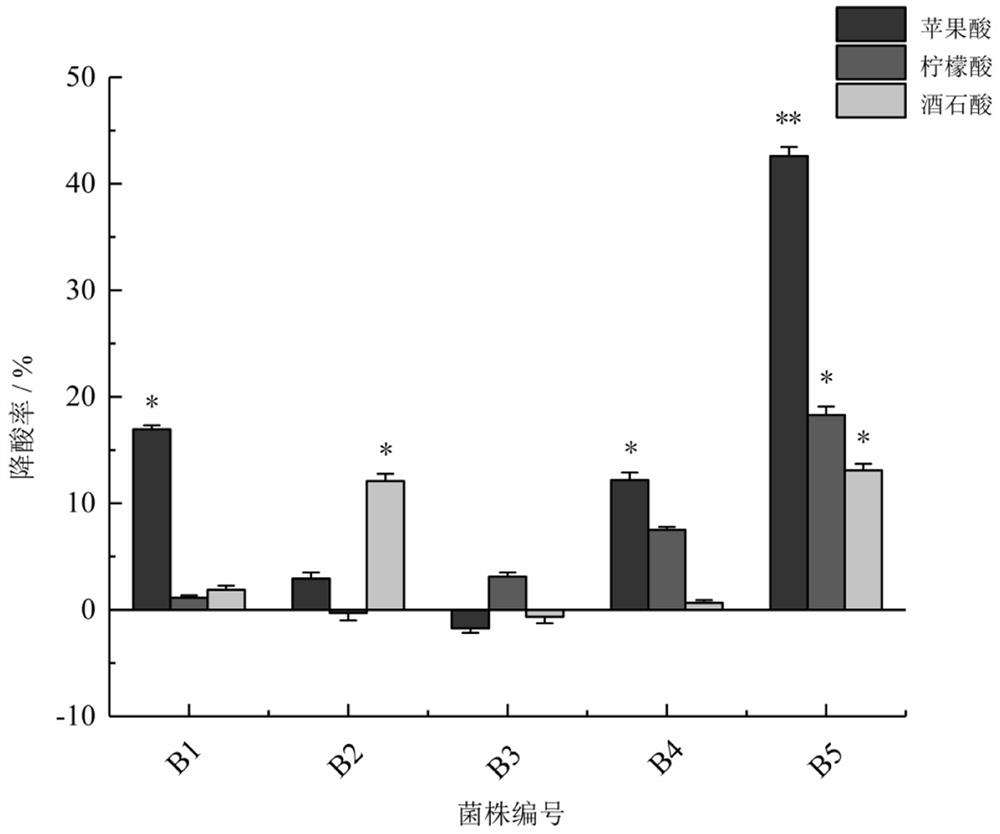
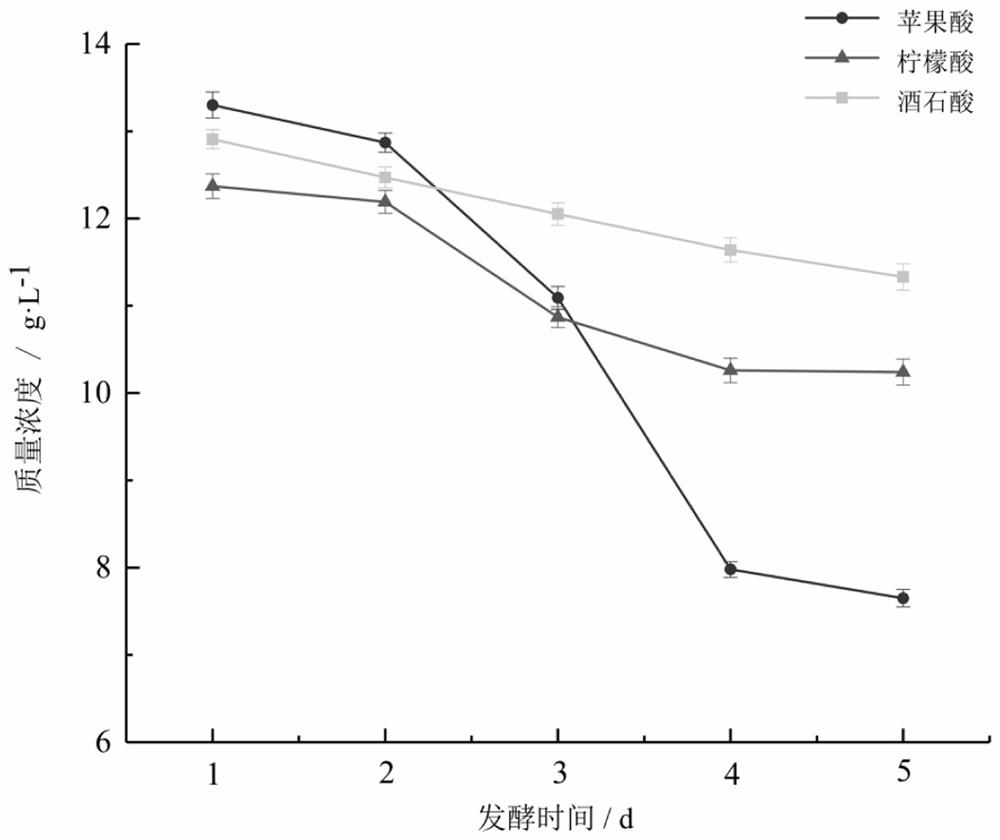
[0059] Example 2 Degradation of Organic Acids in Berries by Dominant Strains
[0060] Determination conditions of high performance liquid chromatography for degrading the organic acids of L. indigo and Seabuckthorn by dominant strains: chromatographic column Agilent ZORBAX Extend-C18 (250 mm × 4.6 mm), gradient elution with methanol-0.1% phosphoric acid aqueous solution (see Table 2), volume The mobile phase with a ratio of 97.5:2.5 was eluted for 10 minutes, and then the methanol phase reached 100% with a short time gradient and equilibrated for 5 minutes, and then the mobile phase was adjusted to 0.1% phosphoric acid solution-methanol volume ratio of 97.5: 2.5, equilibrate for 5 min. The flow rate was 0.7 mL / min, the injection volume was 20 μL, the column temperature was 40 °C, and the detection wavelength was 210 nm. Measure the standard series and samples in turn.
[0061] Draw a standard curve with the concentration of the standard sample versus the peak area, and calcu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com