Five-color FISH probe system and method for detecting multiple genes at one time
A one-time, probe technology, applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc., can solve the problems of missed treatment methods, long waiting time for test results, high cost of testing, etc., to improve Sensitivity and accuracy, shortening the detection time, saving specimens and reagents
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0089] The preparation of embodiment 1 probe
[0090] ① Preparation of the first probe
[0091] (1) Inoculate the BAC strain (RP11-701P18) (Invitrogen, USA) carrying the ALK gene on a fresh LB nutrient agar plate containing antibiotics, and place it at 37°C for overnight culture; the next day, use a 10 microliter white gun First pick the monoclonal strains in good growth state into the liquid medium (2mL LB medium containing antibiotics), shake and culture at 37°C for 6-8h (274rpm); transfer 2mL of the bacteria liquid to 200mL liquid LB medium, Shake at 37° C. (274 rpm), cultivate overnight; follow the operating instructions of QIAGEN’s plasmid mass extraction kit, perform mass extraction and purification of DNA to obtain DNA of the ALK gene.
[0092] (2) For the upstream (5' end) of the ALK gene breakpoint, select the restriction endonuclease HindIII to digest the DNA of the ALK gene to obtain a large fragment of DNA:
Embodiment 2
[0190] The preparation of embodiment 2 probe system
[0191] With formamide buffer (per 2 mL of formamide buffer, 360 μl of 5M NaCl, 40 μl of 1M Tris-HCl, 1 mL of formamide, 2 μl of 10% (w / v) SDS (sodium dodecyl sulfate) , 598 microliters of sterile water, pH7.4) each probe obtained above was diluted respectively to obtain each probe of different concentrations, and the probes of the same gene were classified into a combination, as follows:
[0192] ALK probe combination, including the first, second and third probes (ALK green / orange / blue), the concentrations are: 5ng / uL, 3ng / uL, 20ng / uL respectively;
[0193] ROS1 probe combination, including the fourth and fifth probes (ROS1 green / orange), the concentrations are: 5ng / uL, 20ng / uL;
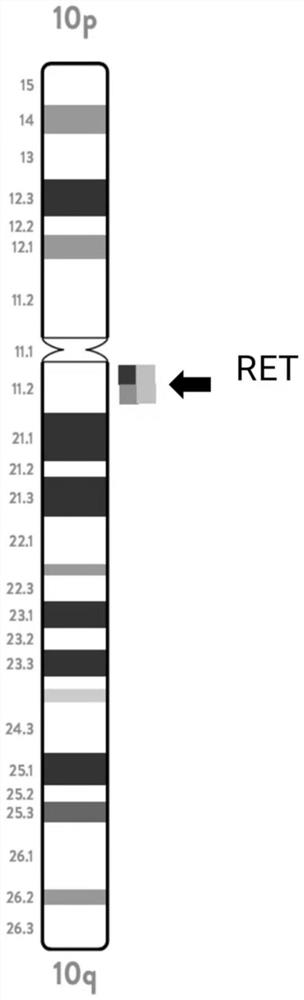
[0194]RET probe combination, including the sixth, seventh, and eighth probes (RET blue / red / gold), the concentrations are: 5ng / uL, 3ng / uL, 20ng / uL;
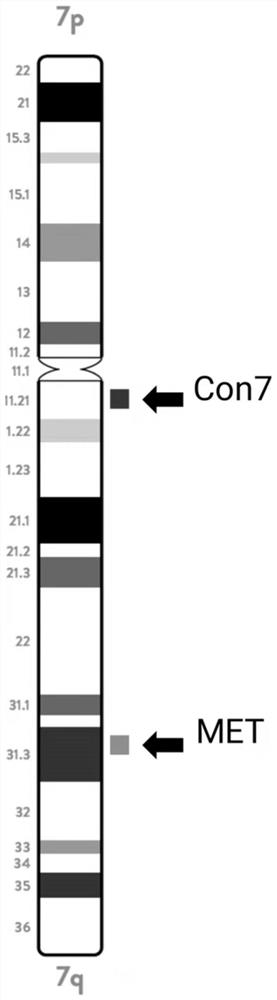
[0195] MET probe combination, including the ninth and tenth probes (MET blue / red), the concentra...
Embodiment 3
[0197] Hybridization reaction of the probe system of embodiment 3 (using the probe system of embodiment 2 to detect the sample)
[0198] 3.1 Sample preprocessing
[0199] ①Slice the sample to be tested at 70°C for 10 minutes.
[0200] ②Xylene (xylene) dewaxing, 10 minutes, twice.
[0201] ③ Incubate slices in 100% ethanol, 100% ethanol, 90% ethanol, and 70% ethanol for 5 minutes respectively.
[0202] ④ Wash with distilled water or deionized water for 2 minutes, twice.
[0203] ⑤ Incubate slices in pretreatment solution preheated to 98°C for 15 minutes.
[0204] ⑥ Immediately transfer the slices to distilled water or deionized water to wash for 2 minutes, twice.
[0205] ⑦ Add protease solution dropwise to the tissue / cell area, and incubate in a constant temperature and humidity chamber at 37°C for about 10 minutes.
[0206] ⑧ Washing buffer for 5 minutes, distilled or deionized water for 1 minute.
[0207] ⑨Dehydration: Dehydrate in 70%, 90%, and 100% ethanol solutions ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com