Tetracycline regulatory protein mutant gene and application thereof in regulatory gene expression and environmental detection
A technology for regulating proteins and tetracyclines, applied in application, genetic engineering, plant genetic improvement and other directions, can solve the problems of insufficient sensitivity, low broad-spectrum, low detection limit of bacterial biosensors, etc., to improve sensitivity, reduce Dox induction concentration, Achieve the effect of in situ detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1 4
[0040] Embodiment 1 The acquisition of tetracycline regulation system gene:
[0041] 1), the acquisition of the wild-type tetracycline regulation system gene:
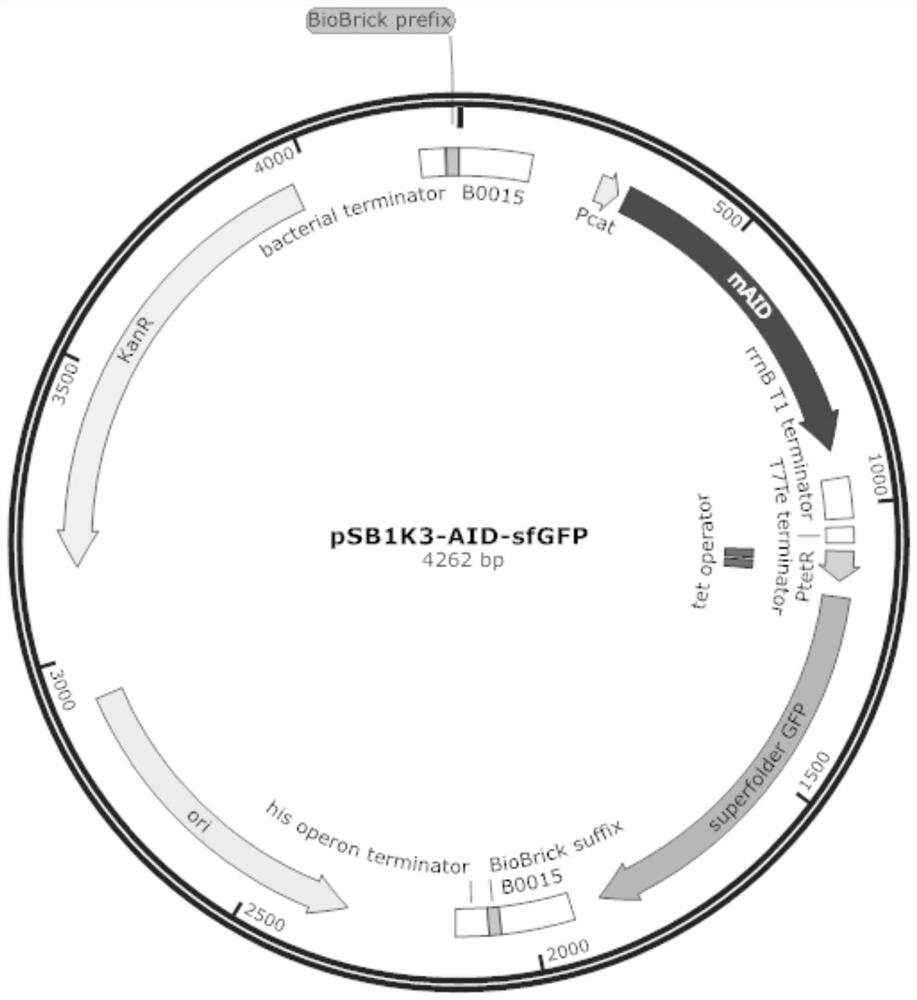
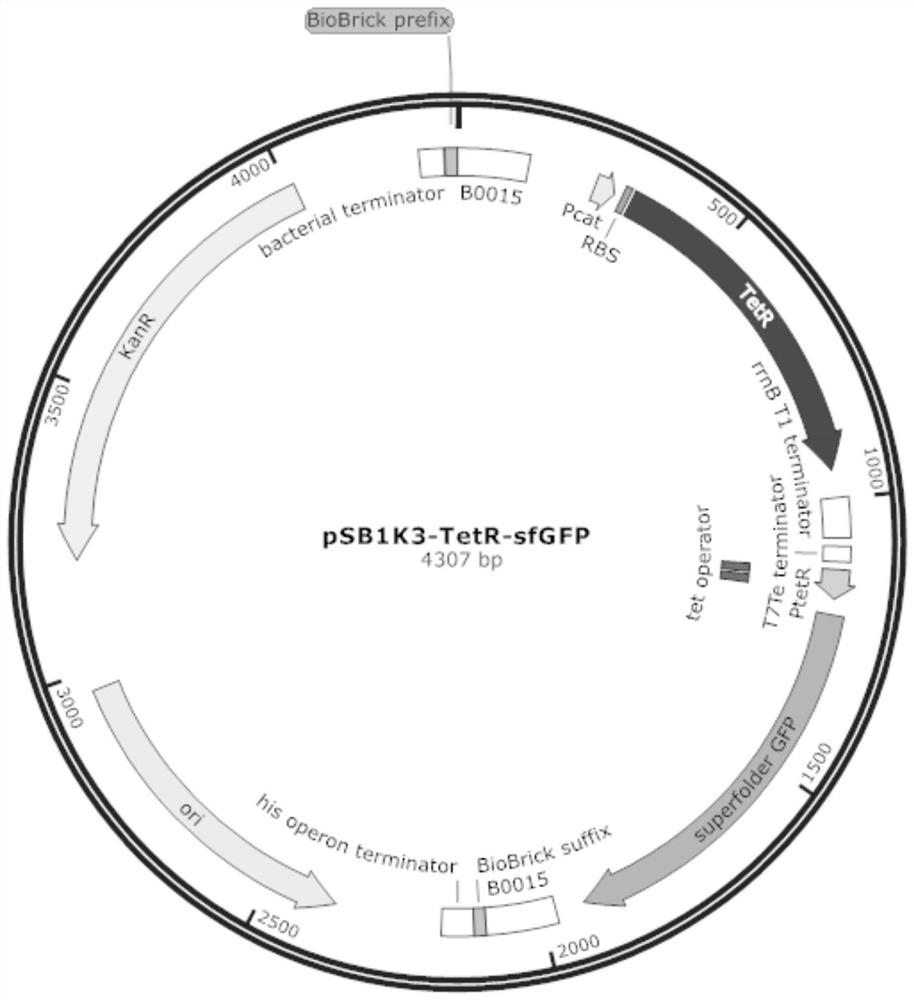
[0042] Using the plasmid pUC57-TetR-sfGFP as a template, PCR amplification was carried out with the primers described in SEQ ID NO: 2 and SEQ ID NO: 3 to obtain the regulatory protein gene TetR, TetR regulatory protein binding site TetO, and an inducible promoter P TetR and a wild-type tetracycline-inducible system for the reporter gene sfGFP,
[0043] 所述质粒pUC57-TetR-sfGFP是根据质粒pFS_0271_pET30_pCat-tetR-Term-ptetA-sfGFP-rrnBterm_Fusicatenibacter_saccharivorans_ARRAY2_FaqI的TetR调控系统部分合成而来的,质粒pFS_0271_pET30_pCat-tetR-Term-ptetA-sfGFP-rrnBterm_Fusicatenibacter_saccharivorans_ARRAY2_FaqI的获得方法参见Schmidt F 2018 paper by et al. (Schmidt, Florian, Mariia Y. Cherepkova Randall J. Platt*. Transcriptional recording by CRISPR spacer acquisition from RNA. Nature 2018; doi:10.1038 / s41586-018-0569-1.).
[0044] Table 1: Primer sequenc...
Embodiment 2
[0060] Dosage Effect Experiment of Evolved Bacterial Biosensors on Dox
[0061] 1) Inoculate the evolved bacterial biosensor on the LB solid medium plate containing kanamycin resistance.
[0062]2) Cultivate overnight at 37°C; at the same time, transform the wild-type tetracycline-inducible recombinant vector pSB1k3-TetR-sfGFP into E.coli DH5α competent cells to obtain wild-type bacterial biosensors, and cultivate wild-type bacterial biosensors simultaneously.
[0063] 3) Pick a single clone, inoculate it in 3 mL of LB liquid medium containing kanamycin resistance, and culture it overnight at 37° C. at 200 rpm to obtain an overnight bacterial liquid.
[0064] 4) Inoculate the overnight bacterial solution from step 2) at 1:20 in fresh LB liquid medium containing kanamycin resistance and expand to logarithmic growth phase (OD 600 =0.4-0.6), to obtain the logarithmic phase bacterial liquid.
[0065] 5) Use deionized water to prepare two sets of tetracycline antibiotic standard ...
Embodiment 3
[0070] Sensitivity experiments of evolved bacterial biosensors
[0071] 1) The evolved bacterial biosensors were inoculated on LB solid medium plates containing kanamycin resistance, cultured overnight at 37°C, and the wild-type bacterial biosensors were simultaneously cultivated.
[0072] 2) Pick a single colony, inoculate it in 3 mL of LB liquid medium containing kanamycin resistance, and culture it overnight at 37° C. and 200 rpm to obtain an overnight bacterial liquid.
[0073] 3) Expand the overnight bacterial solution of step 2) at 1:20 in fresh LB liquid medium containing kanamycin resistance, culture at 37°C, 200rpm shaker to OD 600 =0.4-0.6, to obtain the logarithmic phase bacterial liquid.
[0074] 4) Prepare 200 mg / L Dox stock solution with ultrapure water.
[0075] 5) Add the Dox stock solution to the logarithmic phase bacterial solution obtained in 3), so that the final concentration of the inducer is 200 μg / L, as the induction group; simultaneously take the log...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com