Tumor neoantigen prediction method and neoantigen prediction system based on deep learning model
An antigen, a new technology, applied in the field of biomedicine, can solve the problems of low accuracy and poor efficacy of anti-tumor vaccines, and achieve the effects of high precision, improved effectiveness and reduced cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0118] The present invention provides a neoantigen prediction system, which includes a sample collection device, a transcriptome sequencing data analysis module, and a whole exome sequencing data analysis module (including somatic cell non-synonymous mutation analysis, mutant peptide chain and flanking sequence acquisition, HLA typing analysis unit), neoantigen prediction device.
[0119] The sample collection device includes a tumor cell and a normal cell collection device, and transports the sample to the transcriptome sequencing data analysis module and / or the whole exome sequencing data analysis module, and the tumor cells perform transcriptome sequencing and whole exome sequencing of the sample respectively , normal cells were subjected to whole exome sequencing. The whole exome sequencing data analysis module receives and compares the whole exome sequencing data of tumor cells and normal cells, calculates and presents the somatic cell non-synonymous mutation data to the ...
Embodiment 2
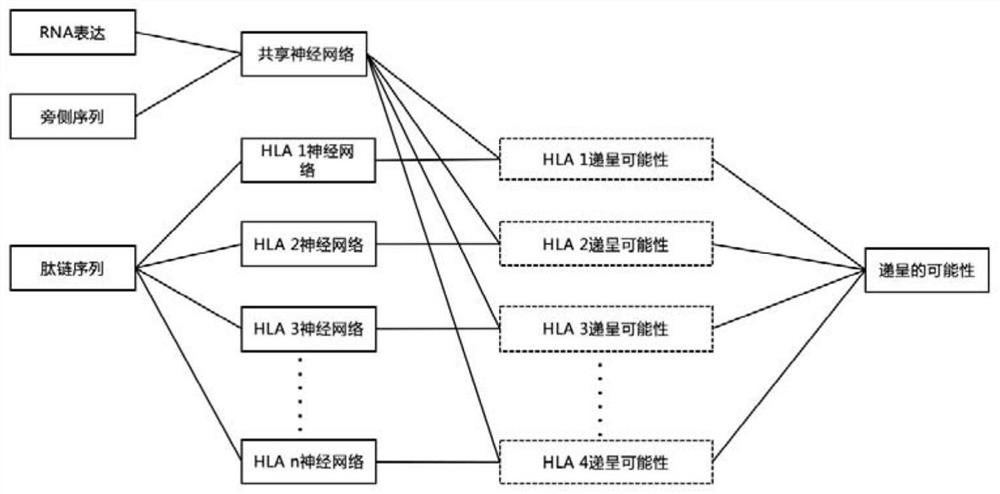
[0127] The construction of the neural network of the neoantigen prediction system is mainly divided into two steps:
[0128] (1) Training data acquisition
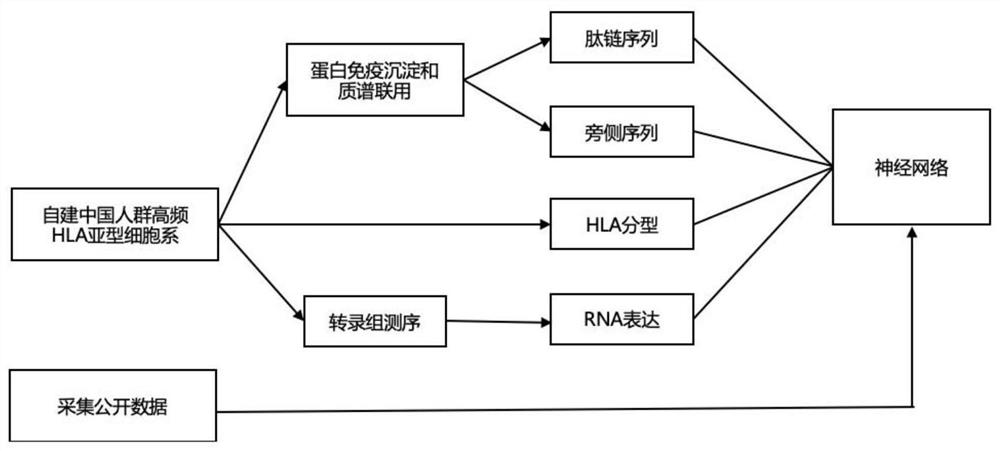
[0129] Our training data acquisition methods are as follows: figure 1 shown. We constructed cell lines with high-frequency HLA subtypes from the Chinese population. Firstly, specific primers for HLA-A, HLA-B and HLA-C were designed, and PCR was used to amplify B-LCL cells ( CRL-2369 TM ) in HLA-A, HLA-B and HLA-C gene fragments, and then these gene fragments were subcloned into retroviral vectors, and finally LCL 721.221 cell line (human HLA class I deletion cell line)( CRL-1855 TM) to obtain HLA subtype cell lines. Use protein immunoprecipitation and mass spectrometry to obtain peptide chain sequences that can bind to a specific HLA molecule, select peptide chains with a length of 8-11 amino acids, and fill in 11 peptide chains with a length of amino acids less than 11, and intercept The 5 amino acids on the lef...
Embodiment 3
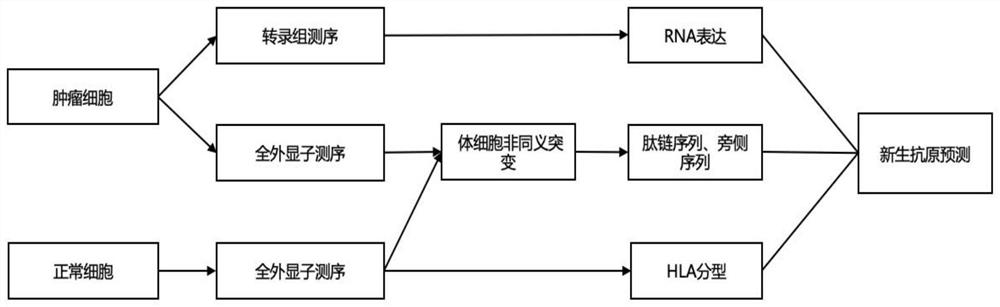
[0133] The process of neoantigen prediction is as follows: image 3 shown. First, collect the patient's tumor tissue and peripheral blood samples, extract DNA, and perform whole-exome sequencing. Firstly, use FastQ software to perform QC (quality control) processing on the sequencing data, and then use BWA software to compare and splicing the sequencing data with the reference genome. , using Mutect2 software to analyze Somatic mutations (somatic mutations) in tumor-blood paired samples, including SNV (single-nucleotide variant, single nucleotide mutation), InDel (insertion / deletion mutation, insertion / deletion mutation), frameshift ( frameshift mutation), etc., screen for non-synonymous mutations, and generate mutant peptide chains and their flanking sequences. The exome sequencing data of peripheral blood samples were analyzed by xHLA software for HLA typing. RNA was extracted from tumor tissue, and transcriptome sequencing was performed to obtain gene expression level (TP...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com