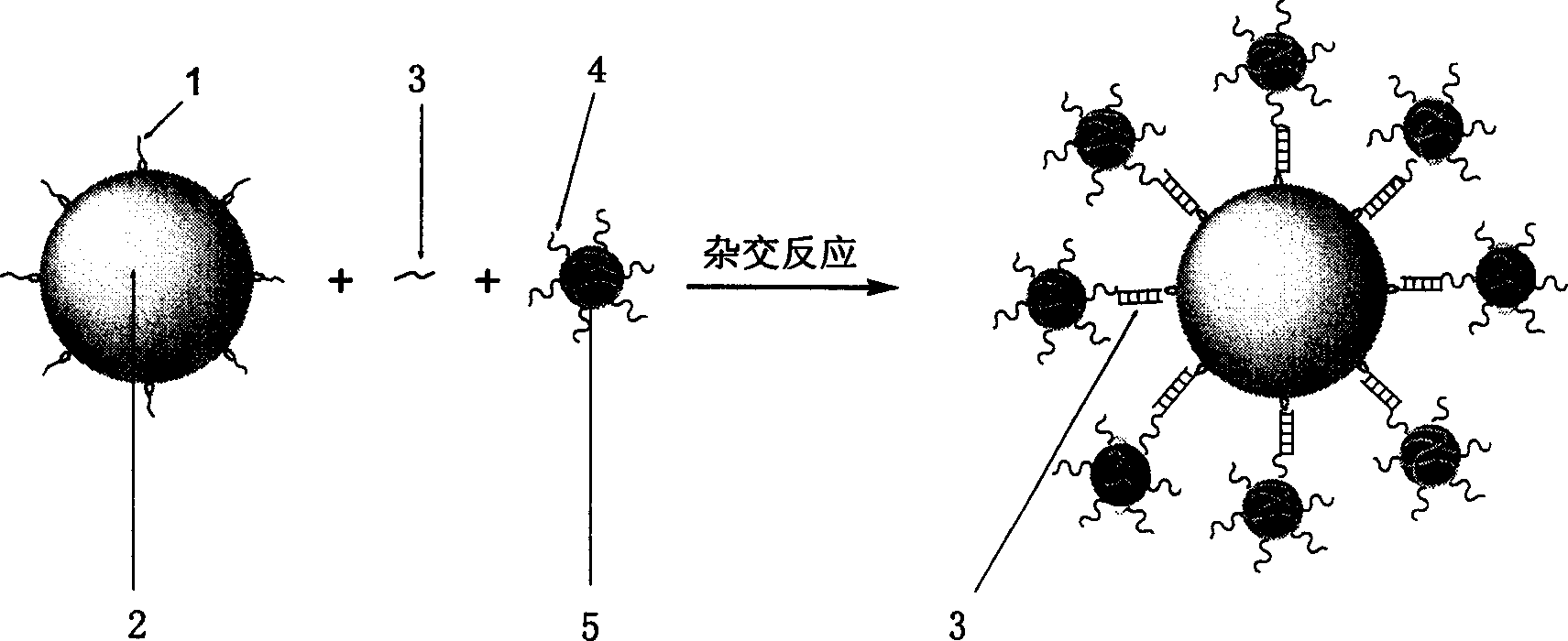
Surface functionalization of gold or silver nanoparticle, and colorimetry detection method for molecule by using the same
A silver nanoparticle and surface functionalization technology, which is applied in biological testing, color/spectral characteristic measurement, material inspection products, etc., can solve the problems of difficult realization of gold nanoparticle labeling, achieve easy purification and quantitative analysis, and avoid detection equipment , the effect of chemical stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0014] Example 1: Determination of DNA by gold nanoparticle-labeled oligonucleotide probes
[0015] Coupling of oligonucleotides to dextran: 0.16ml, 25mg / ml 4-nicotinic acid succinimide ester hydrazine acetone hydrazone (C6-succinimidyl 4-hydrazinonicotinate acetone hydrazone) and 25mg / ml N-succinyl Amine-3-(2-pyridyldithio)-propionate (SPDP, N-succinimidyl3-(2-pyridyldithio)propionate) in anhydrous DMSO solution was added to 4ml, 2mg / ml aminodextran (MW 70KDa ) in 1×PBS solution at room temperature for 2 hours. The resulting dextran conjugate was purified on a Sephadex G25 column using 0.1M NaAc (pH=4.5) as the eluent. The amount of dithio and hydrazone functional groups generated can be quantified with 1,4-dithiothreitol and 4-nitrobenzaldehyde, respectively. Excess aldehyde-modified oligonucleotides were added to a 0.1M NaAc (pH4.5) solution of dextran conjugates and reacted for 12 hours, and the unreacted oligonucleotides were separated and removed with a 30KDa molecular...
Embodiment 2
[0018] Example 2: Silver nanoparticle-labeled oligonucleotide probes measure DNA
[0019]Coupling of oligonucleotides to dextran: 0.16ml, 25mg / ml 4-nicotinic acid succinimide ester hydrazine acetone hydrazone (C6-succinimidyl 4-hydrazinonicotinate acetone hydrazone) and 25mg / ml N-succinyl Amine-3-(2-pyridyldithio)-propionate (SPDP, N-succinimidyl3-(2-pyridyldithio)propionate) in anhydrous DMSO solution was added to 4ml, 2mg / ml aminodextran (MW 70KDa ) in 1×PBS solution at room temperature for 2 hours. The resulting dextran conjugate was purified on a Sephadex G25 column using 0.1M NaAc (pH=4.5) as the eluent. The amount of dithio and hydrazone functional groups generated can be quantified with 1,4-dithiothreitol and 4-nitrobenzaldehyde, respectively. Excessive CHO-modified oligonucleotide probes were added to 0.1M NaAc (pH4.5) solution of dextran conjugates and reacted for 12h, then separated and removed unreacted oligonucleotide probes with a 30KDa molecular weight cut-off ...
Embodiment 3
[0022] Example 3: Gold Nanoparticles Label Biotin Molecules
[0023] Conjugation of biotin to dextran: 5 mg N-succinimide-3-(2-pyridyldithio)-propionate (SPDP) dissolved in 100 μl, 50 mg / ml biotinaminocaproic acid-N- Hydroxysuccinimide ester (Biotinamidohexanic acid N-hydroxysuccinimide ester) anhydrous DMSO solution, this solution was added dropwise to 2.5ml, 2mg / ml aminodextran (MW 70KDa) NaHCO 3 (50mM) solution at room temperature for 2 hours. The resulting dextran conjugates are purified by dialysis in aqueous solution for further use. The amount of biotin on the dextran conjugate can be quantified with HABA (4'-hydroxyazobenzene-2-carboxylic acid).
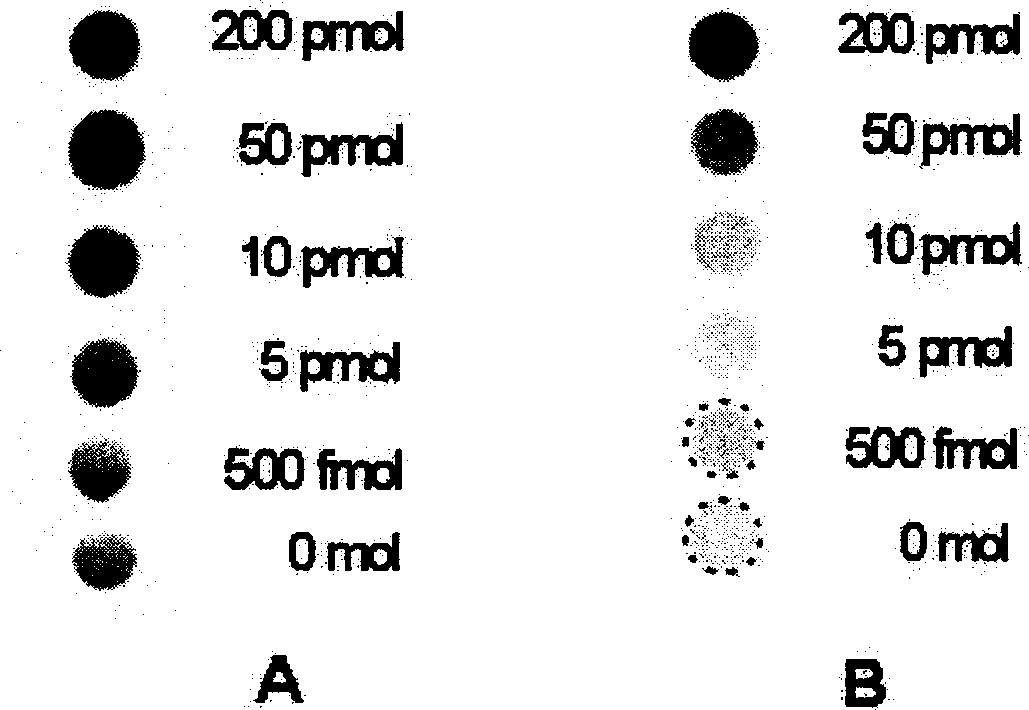
[0024] Preparation of gold nanoparticle-labeled biotin-dextran conjugate probe: add different amounts of biotin-dextran conjugate to a certain amount of gold nanoparticle solution, and record the solution after filtering through a 0.20 μm membrane According to the UV absorption of gold nanoparticles at 520nm, the desired a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com