Protein arginine methyltransferase of Arabidopsis thalianum and its coding gene and application
A technology of arginine methylation and protein, applied in the direction of transferase, application, genetic engineering, etc., can solve the problem that there is no report on histone arginine methylation, achieve high practical application value, and improve tolerance , the effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1, the cloning of the gene AtPRMT5 of Arabidopsis protein arginine methyltransferase
[0036] The obtaining of the full-length cDNA sequence of the gene of Arabidopsis protein arginine methyltransferase comprises the following steps:
[0037] For the human PRMT5 gene sequence (GenBank No.: NM_006109) published in GenBank, a homologous sequence (GenBank No.: NM_119262) was obtained from the Arabidopsis genome by comparison of homologous sequences, and according to the 5' and Design a pair of primers for the 3' end sequence, the primer sequence is as follows:
[0038] P1 (upstream primer); 5'-aagagctccatgccgctcggagagagaggagg-3'
[0039] P2 (downstream primer): 5'-aactcgagaaggccaacccagtacgaacgg-3'
[0040] The total RNA of Arabidopsis leaves was extracted (Promega, SV total RNA isolation system), and the total RNA of Arabidopsis leaves was used as a template to synthesize cDNA with AMV reverse transcriptase (TaKaRa) (according to Plant RT-PCR Kit2.01 (TaKaRa ...
Embodiment 2
[0041] Example 2. Detection of the regulatory effect of AtPRMT5 on the flowering and development of Arabidopsis thaliana
[0042] 1. Screening of AtPRMT5 inactivation homozygous mutants
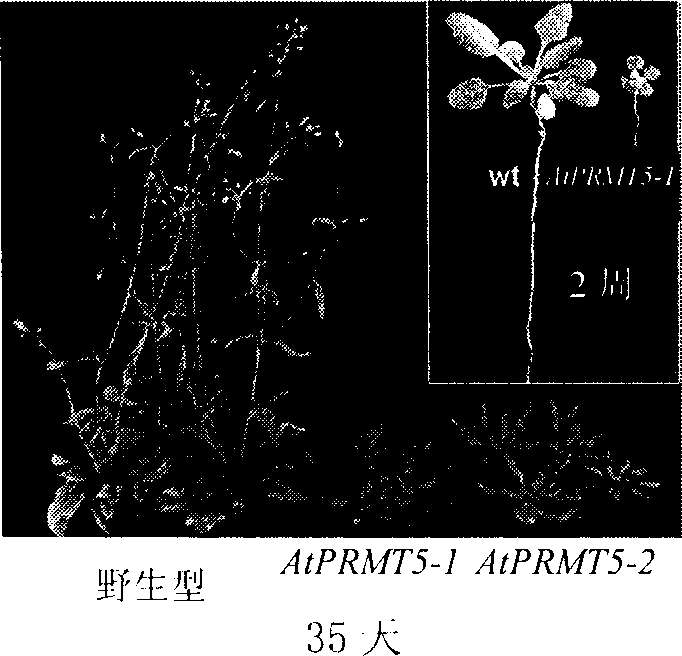
[0043] Live homozygous mutants of Arabidopsis AtPRMT5 were screened by the following method: the seeds (Salk 065814 and Salk 095085) of two different T-DNA insertion mutant lines at the AtPRMT5 site were obtained from ABRC (Arabidopsis Biological Resource Center) (Alonso, J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R., et al. 2003. Genome- wide insertion mutagenesis of Arabidopsisthaliana.Science 301:653-657.), two kinds of AtPRMT5 inactivation homozygous mutants with different T-DNA insertion sites were obtained by PCR identification, named AtPRMT5-1 and AtPRMT5-2 respectively.
[0044] 2. Comparison of the growth of AtPRMT5 inactivation homozygous mutants and wild-type plants
[0045] Harvest the seeds of the AtP...
Embodiment 3
[0051] Example 3. Detection of the influence of AtPRMT5 on histone amino acid modification
[0052] The antibody Anti-H4-R3-sdiMe, which can specifically recognize the symmetrical double-methylation modification of the third arginine position of histone H4 (purchased from Jingmei Bioengineering Company, ABCAM Company) was used to detect the homozygous mutant of AtPRMT5 inactivation and wild-type plants (control, WT) were detected by Western blot, the results were as follows figure 2 As shown, the expression levels of histone methylation in AtPRMT5 inactivation homozygous mutants and wild-type plants are significantly different, indicating that the inactivation of AtPRMT5 leads to symmetrical methylation of the third arginine of histone H4 The level is reduced, affecting the covalent modification of other sites of histone amino acids, changing the codon of histone, thereby affecting the transcription and expression of key genes (such as FLC) in the plant flowering signal transd...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com