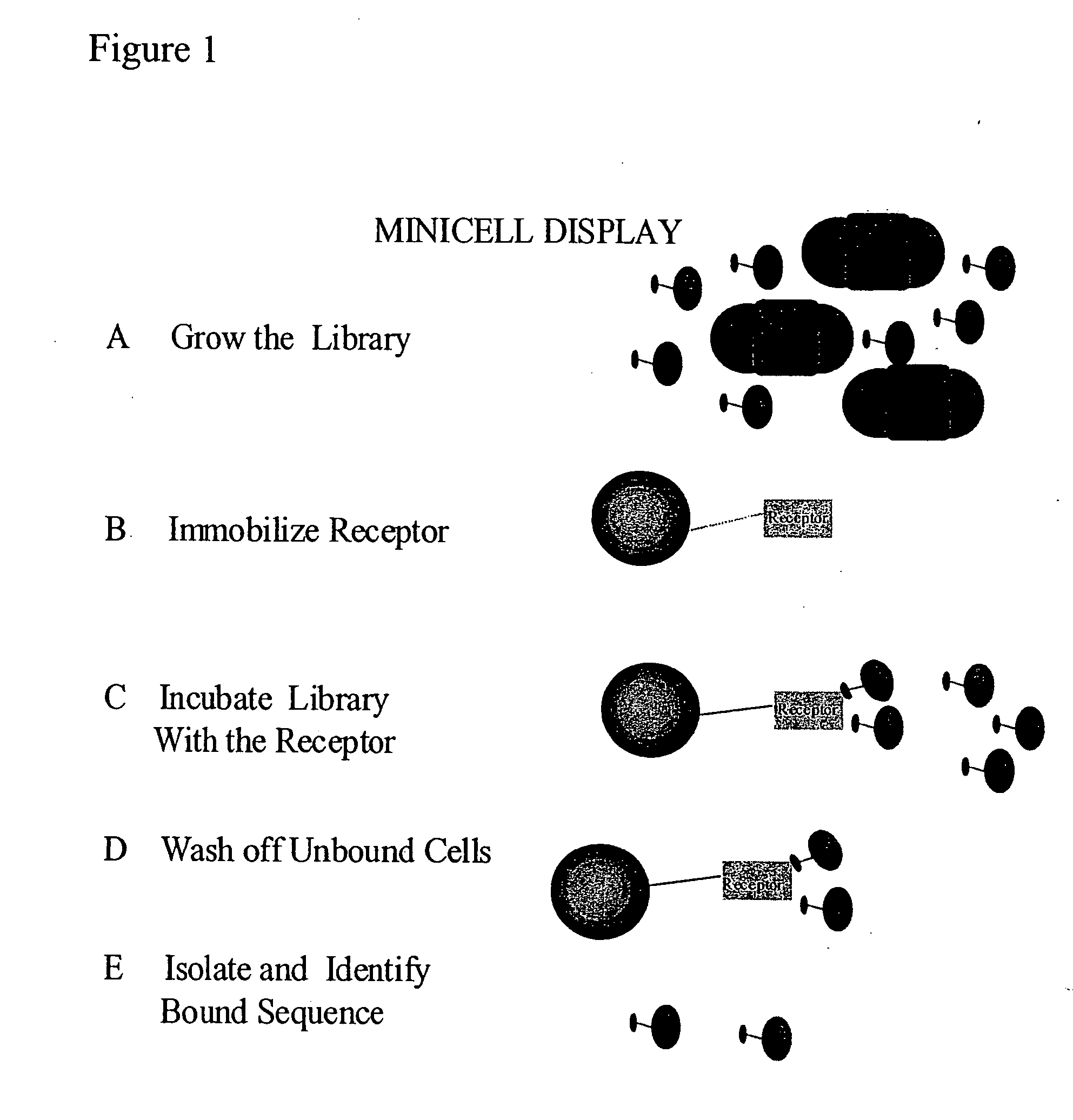
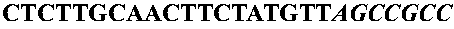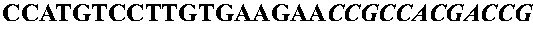Methods to screen peptide display libraries using minicell display
a technology of minicells and peptides, applied in library screening, nucleotide libraries, instruments, etc., can solve the problems of phage assembly, fusion protein cannot reach the cell surface, and constraints affecting the applicability of these systems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of a 17K Antigen Fusion Plasmid for Minicell Display
[0092] A system was constructed to allow the controlled expression of oligonucleotide libraries genetically fused to the 17K antigen of Rickettsia rickettsii. The 17K antigen of R. rickettsii, when cloned into E. coli is displayed to the outer membrane. The N-terminal fragment, containing the lipid modification site, was assembled from the following primers and cloned into pZHA1.3, a plasmid derived from pUC19, by inserting the tac promoter upstream of the unique HindIII site.
[0093] Primers were dissolved in 10 mM Tris, pH 8.5, to a concentration of 100 nmol / μl. 10 μl of each was then mixed, heated to 80° C. for 5 min, cooled to 25° C. (ramp time 1 hour), and incubated at 25° C. for 1 hour. The annealed oligonucleotides were filled in with Klenow, and purified using a QIAquick PCR purification kit (Qiagen), before restriction digestion. The resulting double stranded DNA was cut with XbaI / BamHI and ligated overnight a...
example 2
Construction of the Library
[0095] The primers were synthesized on an Applied Biosystems synthesizer (Forest City, Calif.).
[0096] 1 mM of each primer was separately incubated in 100 μl of 10 mM Tris-HCl buffer, pH 8.0, containing 5 mM MgCl2, 0.5 mM dNTPs and 5 U of Tac polymerase. Reactions were heated to 80° C. for 5 min, cooled to 25° C. (ramp time 1 h), and incubated at 40° C. for 15 min. This procedure was cycled 5 times. After the fifth cycle, 10 μl of each reaction mix was mixed pair-wise with 10 μl of samples from the other reaction as illustrated below. The total volume of each reaction was adjusted to 100 μl with Tris buffer, pH 8.0 containing 5 mM MgCl2, 0.5 mM dNTP, and 5 U tac polymerase and cycled as described above. After the fifth cycle fresh 5 μl of 100 mM dNTP mix was added to each tube and the chaining reaction continued for another 10 cycles.
[0097] The reaction mixes from all 42 tubes (the original 6 primers and the 36 pair-wise tubes) were mixed (see table belo...
example 3
Purification and Labeling of Minicells
[0099] Plasmid pDIP1.0 was transformed into E. coli DS410. Transformed cells were grown in a 250-ml culture to stationary phase (the culture can be grown overnight but must have good aeration). Growing the cells in rich medium minimizes contamination of mini-cells by whole cells. The culture was spun down at 8200× g (7500 rpm in a Sorvall GSA rotor) for 20 minutes at 4° C. The cell pellet was resuspended in 5 ml of the supernatant. Resuspension was very thorough to prevent loss of minicells in the cell pellet during the sucrose gradient step. Pellet was resuspended completely by placing a magnetic stirring bar in the bottom of the centrifuge tube and mixing vigorously for 10 minutes at 4° C. The suspension was carefully layered on a 30-ml sucrose gradient (10-30%). The gradient was centrifuged in a cellulose nitrate ultracentrifuge at 4000× g for 20 minutes at 4° C. (e.g., in an SW27 rotor at 5500 rpm). After centrifugation, a thick, somewhat d...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| total volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com