Urease epitope fusion peptide liposome bacterin for preventing the helicobacter pylori infecting
An anti-Helicobacter pylori and Helicobacter pylori technology, applied in the field of biomedicine, can solve the problem of not completely preventing Helicobacter pylori colonization and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
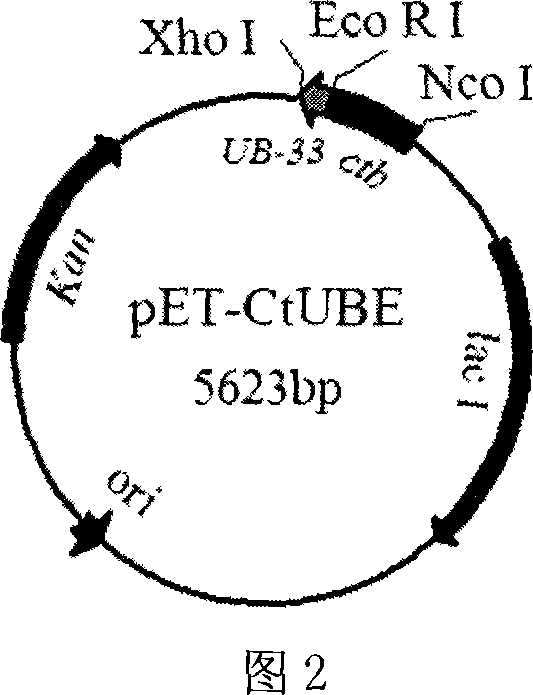
[0039] Example 1 Construction of Fusion Peptide CtUBE Expression Vector
[0040] (1) Design and synthesize PCR primers and introduce NcoI and EcoRI sites at the same time:
[0041] PCTB01: 5′-AAAAA CCATGG GCACACCTCAAAATATTACTGATTTG-3′,
[0042] PCTB02: 5′-AA GAATTC GCCGCCATTTGCCATACTAATTGCGGCAATCGCAT-3';
[0043] (2) Using the Vibrio cholerae genomic DNA as a template, the CTB coding sequence was amplified by PCR (94°C pre-denaturation for 3 minutes; 94°C for 45s, 56°C for 50s, 72°C for 50s, 40 cycles; 72°C extension for 10min) to amplify the CTB coding sequence 1);
[0044] (3) The PCR product was double digested with NcoI and EcoRI and cloned into the multiple cloning site of pET-28a to obtain the intermediate vector pET-CtUBE-Pre, which was transformed into Escherichia coli DH5α for amplification;
[0045] (4) Synthesize the UreB epitope coding sequence and introduce EciRI and XhoI sites simultaneously:
[0046] UBP1: 5′- AATTC TGCCACCACTTGGATAAAAGCATTAAAGAAGATGTTC...
Embodiment 2
[0051] Example 2 Establishment of fusion peptide CtUBE expression, separation, renaturation and purification system
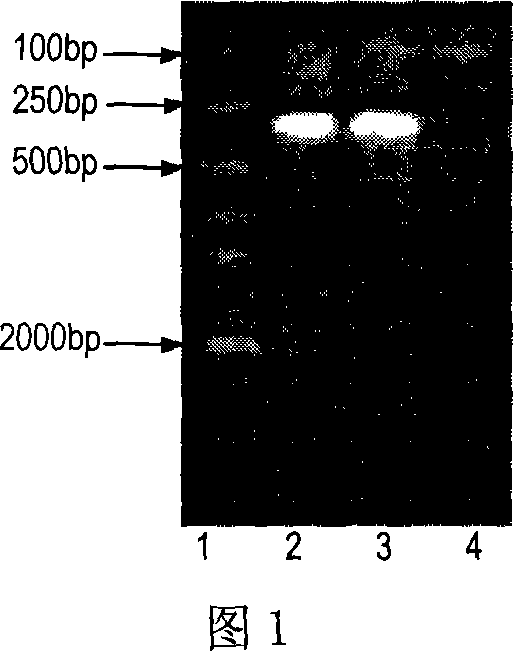
[0052] (1) Transfer the positive transformants with correct sequencing into LB medium, induce expression with 1mM IPTG (accompanying drawing 4), analyze the expression form of the fusion protein through 12% SDS-PAGE, and analyze the expression amount with electronic scanning simultaneously;
[0053] (2) The fusion peptide is expressed in the form of inclusion bodies. After expanding the expression culture, the bacterial cells are lysed with a lysate containing lysozyme, and the inclusion bodies are collected by centrifugation;
[0054] (3) After the inclusion bodies were washed 5 times with Wash Buffer, they were dissolved with inclusion body dissolution buffer, and about 10 times the volume of Renaturation Buffer was slowly added, and transferred to 4°C for renaturation for more than 48 hours;
[0055] (4) After the refolded fusion protein is fully dialyzed wi...
Embodiment 3
[0058] Example 3 Preparation of fusion peptide CtUBE liposome vaccine
[0059] (1) Add 0.314g lecithin and 0.126g cholesterol respectively in eggplant-shaped bottle, and add 6ml chloroform to make lecithin and cholesterol dissolve and mix;
[0060] (2) Evaporate chloroform to dryness on the rotary evaporator, add 2.26ml chloroform and 3.74ml ether, dissolve lecithin and cholesterol film;
[0061] (3) Add 2ml of 1mg / ml CtUBE solution (dissolved in PBS with pH 8.04), and sonicate in ice bath for 4min;
[0062] (4) Reduce the pressure on the rotary evaporator to about 0.05MPa, and evaporate the organic solvent at 37°C;
[0063] (5) Add 10ml of PBS (pH8.04) to the eggplant-shaped bottle and rotate at high speed for 3-4h to obtain a light milky yellow liposome suspension;
[0064] (6) Regulate the liposome particle size with 1.0 μm, 0.8 μm and 0.4 μm microporous membrane successively;
[0065] (7) Take 1ml of liposome suspension and put it into a 1.5ml centrifuge tube, centrifug...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com