High throughput real-time detection technology for drug resistance gene frequency of Sclerotinia sclerotiorum population
A technology for gene frequency and real-time detection, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., to achieve the effect of improving specificity, improving amplification efficiency, and simple detection and monitoring
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Example 1. Screening of specific primers suitable for real-time quantitative PCR (Real-time quantitative PCR).
[0067] The mutation of amino acid 198 of β-tubulin gene from glutamic acid (Glu) to alanine (Ala) is the main cause of field resistance of S. sclerotiorum to carbendazim, and the corresponding code According to this mutation, the upstream ASO-specific primer 5'-TGGTCGAGAACTCTGACGC-3', which can amplify the characteristic band of resistant S. sclerotiorum, was designed. This primer cannot be used for real-time quantitative PCR detection. Because there is a secondary structure in the amplification product; since SYBR Green I is a fluorescent dye that only binds to the DNA double strand, but it cannot select a specific DNA template, the quantification of this method requires that the primers cannot form dimers. No mismatch occurs. In addition, in the present invention, compared with the sensitive strain, only one base has changed in the β-tubulin gene sequence ...
Embodiment 2
[0095] Example 2. SYBR Green I fluorescent dye quantitative PCR system optimization
[0096] 2.1 Annealing temperature In order to ensure the stability and reliability of the detection, the experiment selected Bao Biological Engineering (Dalian) Co., Ltd. Premix Ex Taq TM (Perfect Real Time) DRR041S kit. Enzyme, Mg in the kit 2+ , dNTP, etc. have been optimized, only need to add your own template and primers, therefore, the experiment is only optimized for annealing temperature and primer concentration.
[0097] The annealing temperature Tm value is an important guarantee for the specificity of the reaction. If the annealing temperature is too low, non-specific amplification will occur. Therefore, choosing a higher annealing temperature can greatly reduce the non-specific binding between the primer and the template and improve the specificity of the PCR reaction. However, too high annealing temperature will reduce the amplification efficiency, so it needs to be optimized. ...
Embodiment 3
[0100] Example 3 The sensitivity of SYBR Green I fluorescent dye quantitative PCR
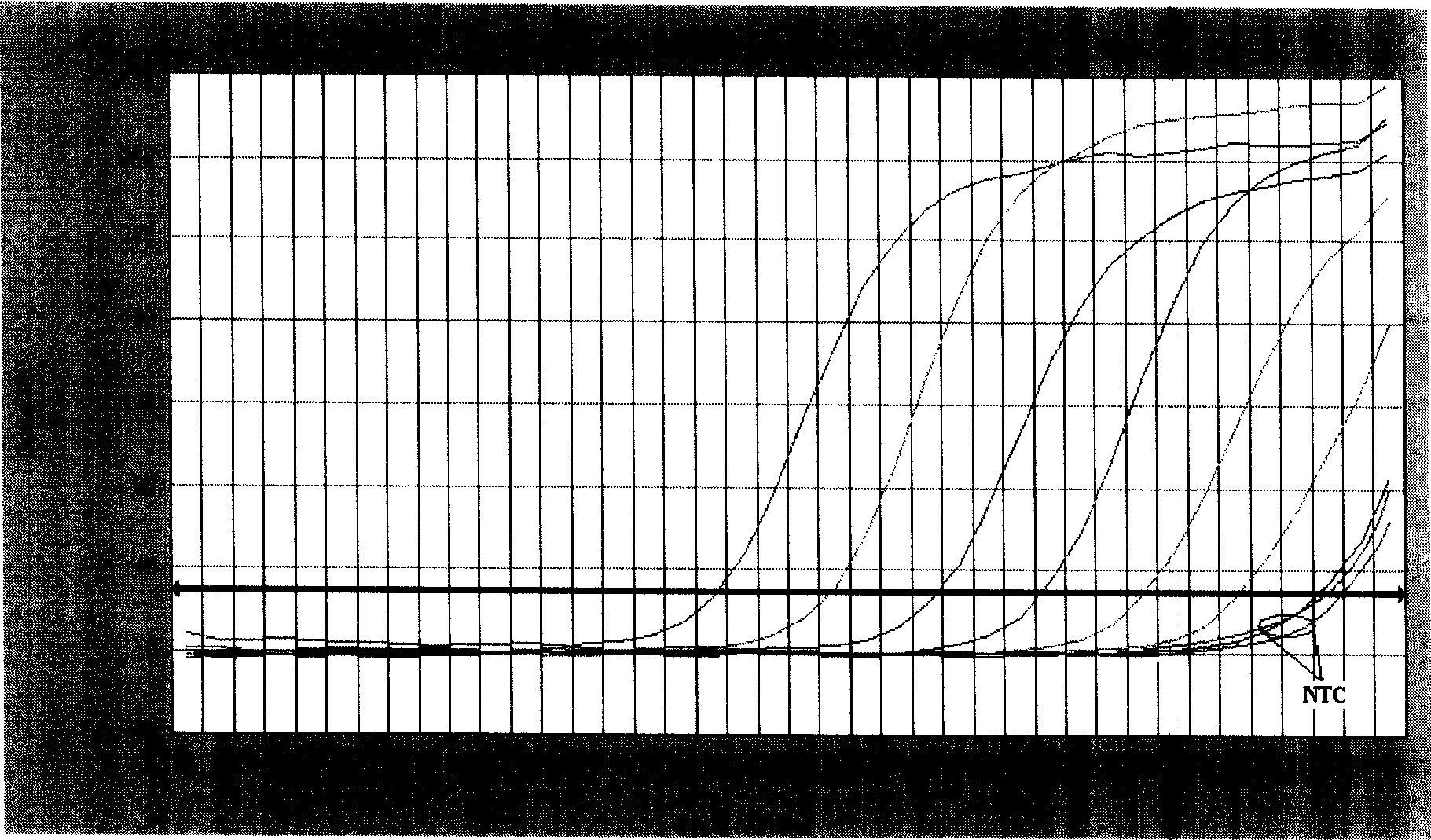
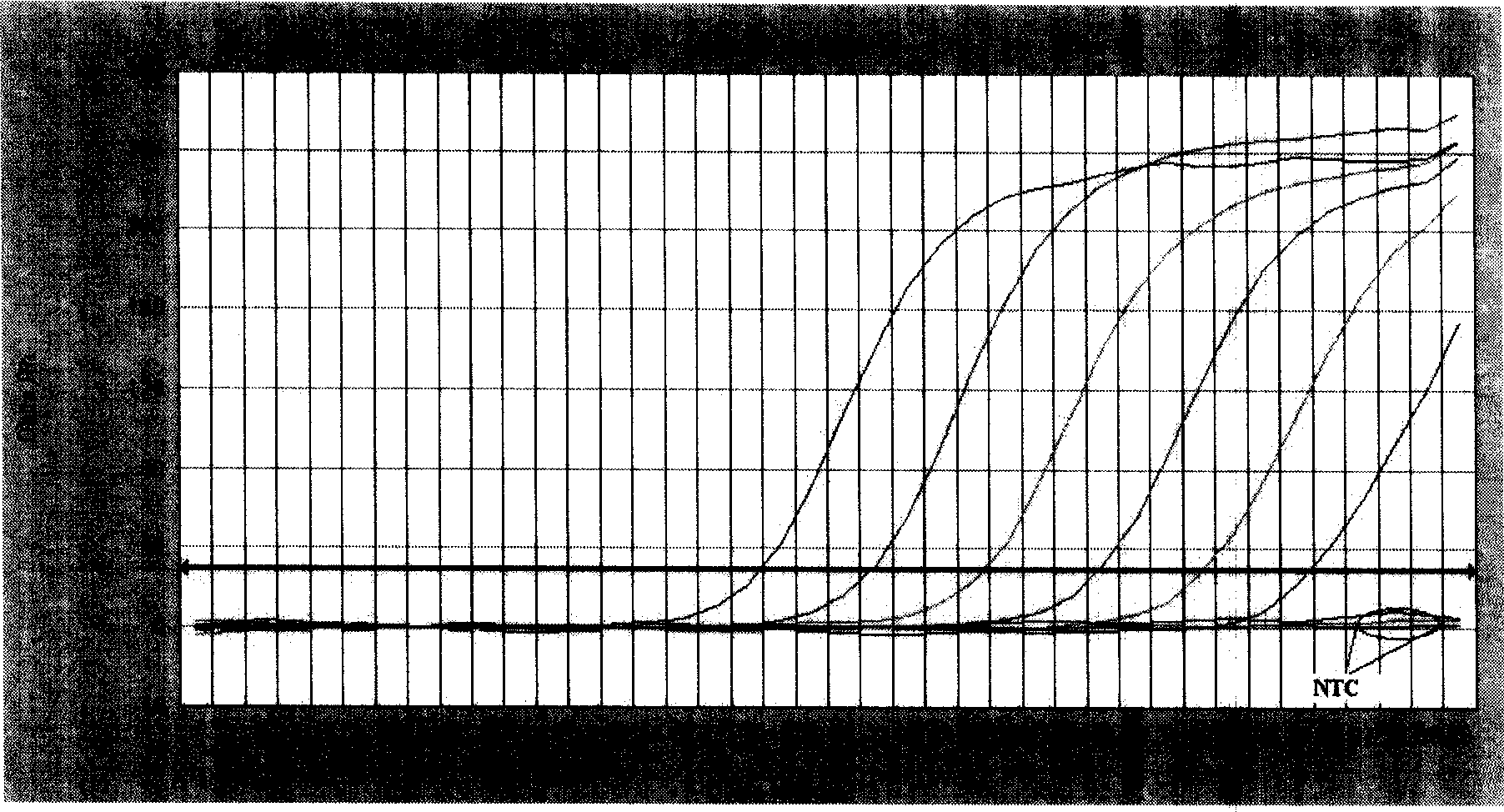
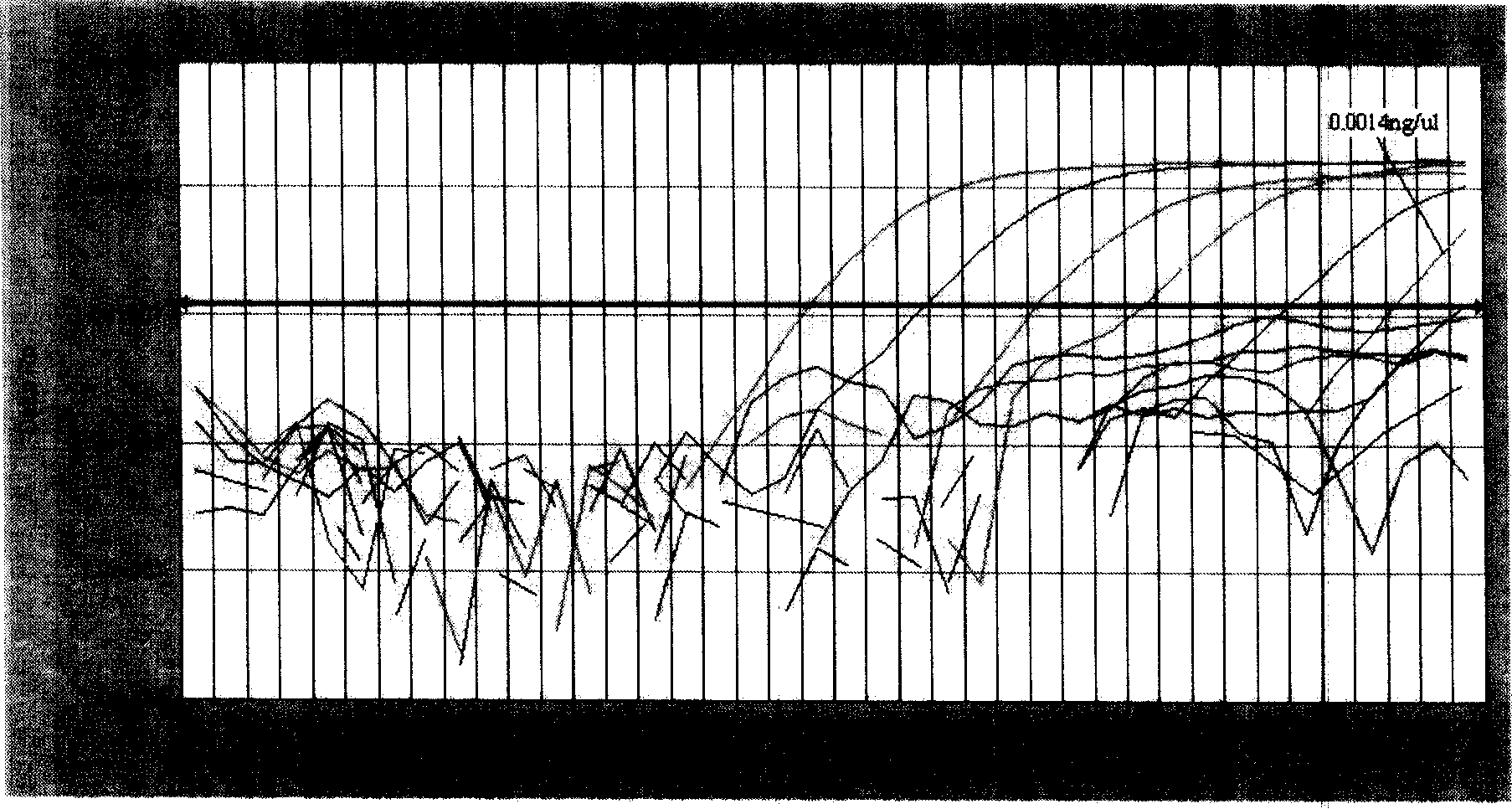
[0101] Standards of strain JSJ6 genomic DNA were serially diluted into 10 gradients: 140ng / μL, 14ng / μL, 1.4ng / μL, 1.4×10 -1 ng / μL, 1.4×10 -2 ng / μL, 1.4×10 -3 ng / μL, 1.4×10 -4 ng / μL, 1.4×10 -5 ng / μL, 1.4×10 -6 ng / μL, 1.4×10 -7 ng / μL, quantitatively amplified with two pairs of primers respectively; the obtained amplification pattern ( image 3 ), when the template concentration is less than 1.4×10 -3 After ng / μL, the fluorescence value of the amplification curve is lower than the threshold value; while the product is detected by 1.5% agarose gel electrophoresis, when the template concentration is less than 1.4×10 -2 No electrophoresis band after ng / μL ( Figure 4 ). It can be seen that the lowest sample concentration detectable by SYBR Green I fluorescent dye quantitative PCR is 1.4×10 -3 ng / μL (2.8×10 per 25μL system -3 ng, 3.17×10 3 copy), the sensitivity is at least 10 to 100 times...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com