Sucrose isomerase gene and high-efficiency expression method thereof
A sucrose isomerase and high-efficiency expression technology is applied in the field of sucrose isomerase gene and its high-efficiency expression, and achieves the effects of high-efficiency expression, short induction time, convenient use and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Extraction of total DNA from Erwinia rhapontici NX-5.
[0032]E.rhapontici NX-5 strain (CCTCC NO: 2222) was cultured in SB liquid medium (peptone 10g / L, yeast extract 5g / L, NaCl 10g / L) for 12h, collected by centrifugation at 8000r / min, sterile water Wash, collect and suspend the precipitate in 500 μL Tris-EDTA (trishydroxymethylaminomethane-ethylenediaminetetraacetic acid) buffer, add 5 μL RNase, incubate at 37 °C for 30 min, add 30 μL 10% SDS (sodium dodecyl sulfate) and 15 μL of proteinase K, incubated at 37°C for 60 min, added 100 μL of 5M NaCl solution and 80 μL of CTAB (cetyltrimethylammonium bromide), incubated at 65°C for 20 min, and added 700 μL of phenol: chloroform: isoamyl alcohol Extract the mixed solution with a ratio of 25:24:1, centrifuge at 10,000 r / min, extract the supernatant with 700 μL of chloroform:isoamyl alcohol with a volume ratio of 24:1, centrifuge at 10,000 r / min, and centrifuge the supernatant with 1,400 μL of ice isoamyl alcohol ...
Embodiment 2
[0033] Example 2: Cloning of the gene encoding SIase.
[0034] The E.rhapontici NX-5 total DNA obtained in Example 1 is used as a template, and the following nucleotide sequences are used as primers:
[0035] Primer 1: TT AAGCTT CCATGG ATTTCTCAAGGATT, respectively introduced HindIII, NcoI restriction sites (underlined part).
[0036] Primer 2: GTAAATATTTTGAATTAGGC GAGCTC CCTAGG TT, respectively introduce BamH I and Xho I double restriction sites (underlined part).
[0037] The PCR reaction was carried out in a 20 μL system: 2 μL of 10×PCR buffer, 2 μL of 2.5 mmol / LdNTP, 2 μL of 10 μmol / L primer 1 and primer 2, 2 μL of template DNA, 25 mmol / L MgCl 2 2 μL, TaqDNA polymerase 1 μL, add double distilled water to 20 μL.
[0038] PCR reaction conditions: Denaturation at 95°C for 3 minutes, and then cycle, (94°C for 30s; 45°C for 30s; 72°C for 2min; 72°C for 5min), a total of 25 cycles, amplified to obtain a 1800bp PCR fragment, cut the gel and recover, and recover the frag...
Embodiment 3
[0039] Example 3: Construction of pal I gene on expression vector.
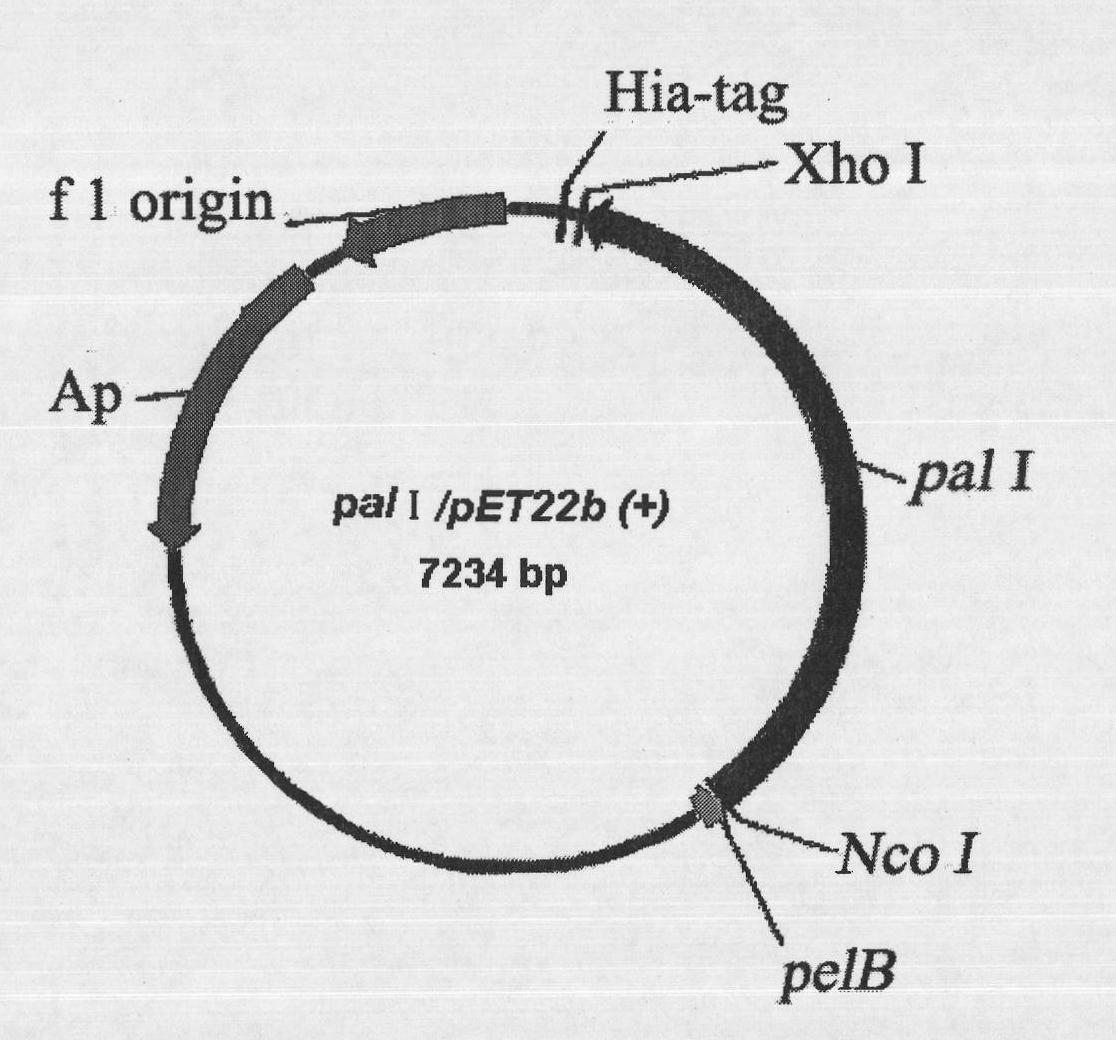
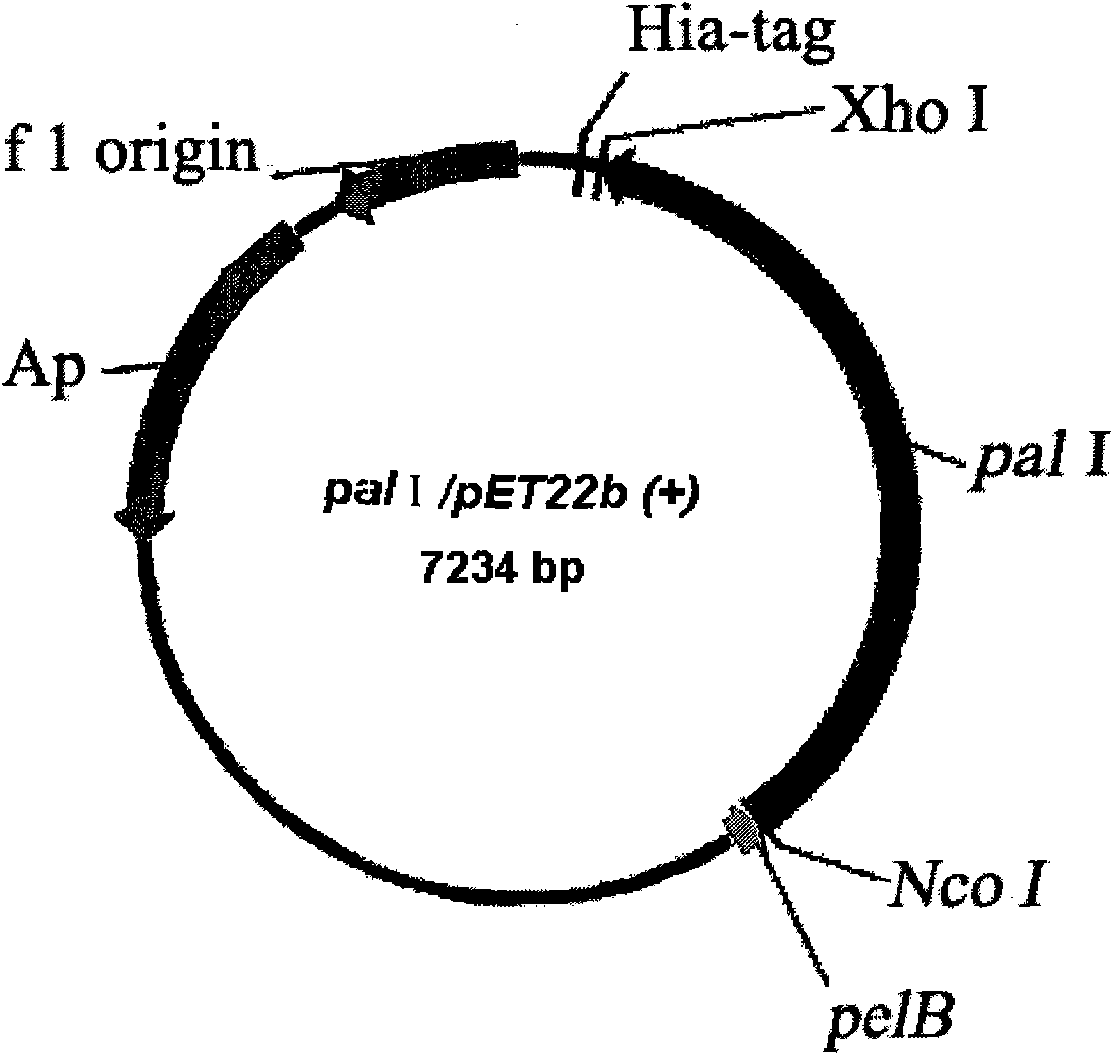
[0040] The plasmid used to construct the expression vector is pET22b(+), with pelB signal peptide and His-tag marker. The pET22b(+) plasmid and pal I gene were digested with Xho I and Nco I double enzymes, the gel was cut to recover pal I, and then ligated with T4 ligase overnight at 16°C. The ligated product was transformed into E.coli BL21 (DE3) competent cells, and the Cultivate overnight at 37°C, select transformants and carry out liquid culture in LB of 100 μg / mL ampicillin, and then extract the plasmid to obtain the enriched pal I / pET22b(+) plasmid ( figure 1 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com