Thermus siphoviridae phage 4 (TSP4) DNA helicase and polynucleotide coding same
A DNA helicase and polynucleotide technology, applied in the biological field, can solve the problems of poor stability and heat resistance, affecting the efficiency of PCR reaction, etc., so as to improve the efficiency, improve the melting efficiency, and improve the amplification efficiency. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
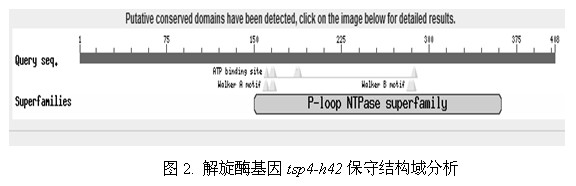
[0031] Example 1: Cloning and expression of a DNA helicase
[0032] 1. Amplification of TSP4 phage DNA helicase gene (using phage TSP4DNA as template)
[0033] (1) The primer sequences used for amplification of TSP4 phage DNA helicase gene are as follows:
[0034] Forward primer: 5'- CATATG ACGGACATAAAGCTGGAG -3'
[0035] Reverse primer: 5'- CTCGAG GGTAAGAGAGAAATCAAAGTT -3'
[0036] (2) The amplification system is as follows:
[0037] Table 1: Amplification reaction system components
[0038] template 10ng Premix exTaq 25 μl Primer 1 μl each Double distilled water ddH 2 o Replenish to 50 μl
[0039] (3) The amplification conditions are as follows:
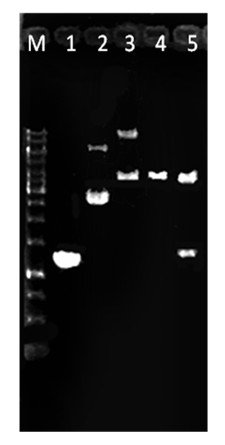
[0040] Mix the reaction system evenly, pre-denature at 94°C for 4 min, then denature at 94°C for 30 s, anneal at 60°C for 30 s, extend at 72°C for 30 s, and after 30 cycles, extend at 72°C for 10 min. After the reaction, 5 μl of the product was taken and analyzed by electrophoresis in...
Embodiment 2
[0118] Embodiment 2 Determination of the ATPase Enzyme Activity of Recombinant Helicase Protein
[0119] The method of ATPase enzyme activity is determined by malachite green ammonium molybdate method. When ATP is hydrolyzed into ADP by helicase and free phosphate is released, blue complexes can be produced with molybdate. By measuring the absorbance, it can be determined Its ATPase enzymatic activity, the method is as follows:
[0120] The preparation method of malachite green dye stock solution was as follows: 60 mL of concentrated sulfuric acid (1.84 g / mL) was slowly added to 300 mL of H 2 O, cooled to room temperature, and added 0.44 g malachite green dye.
[0121] The sample is: 1 for buffer + dsDNA + ATP + Mg 2+ (blank control), 2 is recombinant tsp4-h42 helicase + ATP, 3 is recombinant tsp4-h42 helicase + dsDNA + ATP, 4 is recombinant tsp4-h42 helicase + dsDNA + ATP + Mg 2+ .
[0122] The reaction mixture (200 μl) for the determination of ATPase activity contains ...
example 3
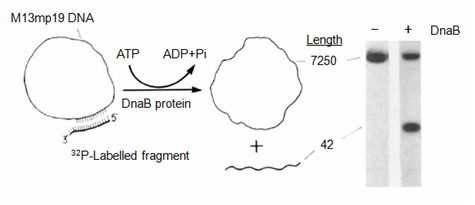
[0125] Example 3 Measurement of helicase activity by isotope labeling
[0126] 1. For the measurement principle see figure 1 .
[0127] 2. Detwisted template preparation
[0128] a. Using T4 polynucleotide kinase (Takara) and [γ- 32 P] ATP marks the 3' sequence of the synthetic primer sequence p42, and the p42 primer sequence is:
[0129] 5'-CAGTGAATTCGAGCTCGGTACCCGGGGATCCTCTAGAGTCGA
[0130] b. The p42 labeled with the radioactive isotope is annealed with the genome of the M13 phage to form a partial double strand.
[0131] c. Annealing to form a labeled unwrapped template:
[0132] The annealing process is as follows: boil the corresponding primer and template in the annealing buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl) at a ratio of 1:1.5 for 3 min, and naturally cool to room temperature, usually 4 h, to form End-labeled DNA p42 / M13.
[0133] 3. Determination of helicase activity
[0134] The standard helicase activity assay system is 20 μl, and the reaction mixt...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com