Cold-adapted marine yeast BoHai Sea-9145 low-temperature alkaline lipase gene, amino acid sequence and recombinant lipase
A lipase gene, marine yeast technology, applied in genetic engineering, plant genetic improvement, fermentation, etc., can solve the problems of complex metabolites, low occupancy, cumbersome extraction and purification steps, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0146] Obtaining of embodiment 1 lipase gene conservative sequence
[0147] The inventor screened a cold-adapted marine yeast Bohai Sea-9145 that produces low-temperature alkaline lipase, which belongs to a subspecies of Yarrowia lipolytica (patent authorization number ZL200410096668.7); Cloning of the lipase gene.
[0148] 11 Acquisition of Conserved Sequence of Marine Low-temperature Lipase Gene
[0149] The degenerate primers F1 and R1 were designed according to the conserved sequences of lipase genes of different species and the codon preference of Y. lipolytica. The sequences are as follows
[0150] F1 5'-GTSTTYMGAGGHACYTT-3'
[0151] R1 5'-CC SARRGAGTGDCCRGT-3'
[0152] The total DNA of the screened wild cold-adaptive marine yeast Bohai Sea-9145 strain was used as a template for PCR amplification under the following conditions:
[0153] Reaction conditions: pre-denaturation at 94°C for 5 minutes; denaturation temperature at 94°C for 1 minute; annealing temperature at...
Embodiment 2
[0164] The preparation of embodiment 2 recombinant lipase
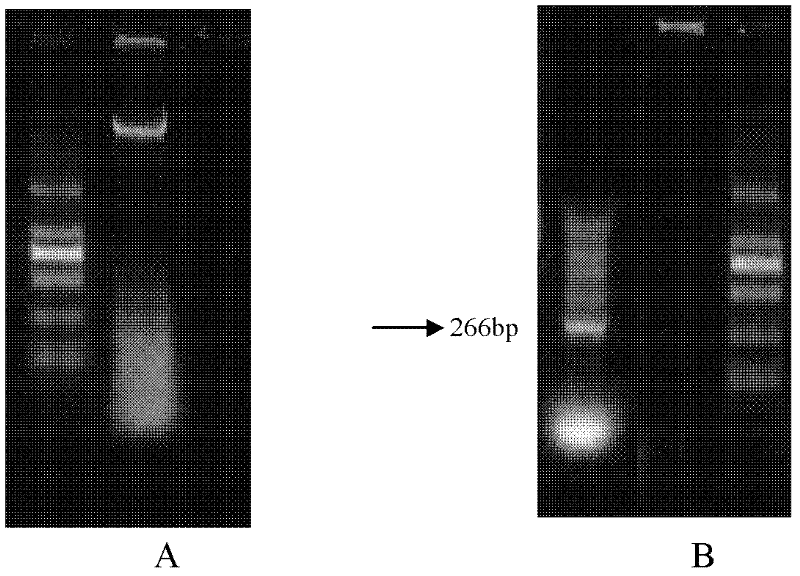
[0165] 2.1 Cloning of lipase LipY gene
[0166] According to the marine low-temperature alkaline lipase gene sequence, two expression primers F2 and R2 were designed,
[0167] F2 5'-GAC GAATTC
[0168] GCAGGCGTGTCTCAAGG-3'
[0169] R2 5'-AAC GCGGCCGC GTTCTCAACTTGTGGGG-3'
[0170] The underlined marks are EcoRI and NotI restriction sites respectively, and a base encoding 6×histidine (indicated in italics) is added to the 5' end of the primer F2.
[0171] The genomic DNA of the cold-adapted marine yeast Bohai Sea-9145 strain was used as a template, and F2 and R2 were used as primers for PCR amplification. The conditions are as follows:
[0172] Reaction conditions: pre-denaturation at 94°C for 5 minutes; denaturation temperature at 94°C for 1 minute; annealing temperature at 54°C for 1 minute; extension temperature at 72°C for 1 minute; after 30 cycles, extension at 72°C for 10 minutes.
[0173] The PCR produ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com