Alkaline pectinase poly lactic acid (PLA) and gene and application thereof
A pectinase and alkaline technology, applied in the field of genetic engineering, can solve the problems of low expression of alkaline pectinase and not being widely used
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0094] The cloning of embodiment 1 Klebsiella pectinase coding gene plA
[0095] Klebsiella sp. is derived from the soil samples of Guangxi fruit juice factory, and the genomic DNA of Klebsiella sp. is extracted: centrifuge the bacterial liquid cultured in LB medium at 30°C for 2 days at 10,000rpm for 10min , Weigh 50 mg of bacteria sludge and wash with 500 μL of sterile water, and centrifuge to get the precipitated bacteria. Resuspend the precipitated bacteria in 500 μL of lysozyme mixture, incubate at 37°C for 1 hour, add 100 μL of enzyme solution and continue to incubate at 45°C for 30 minutes until the bacteria solution is transparent, add 10% SDS to a final concentration of 2%, and stir for about 5 minutes Until the viscosity of the bacterial liquid drops significantly, centrifuge at 15,000 rpm for 10 minutes to remove debris. The supernatant was extracted with equal volumes of phenol, phenol:chloroform, and chloroform in sequence. Take the upper layer solution and add ...
Embodiment 2
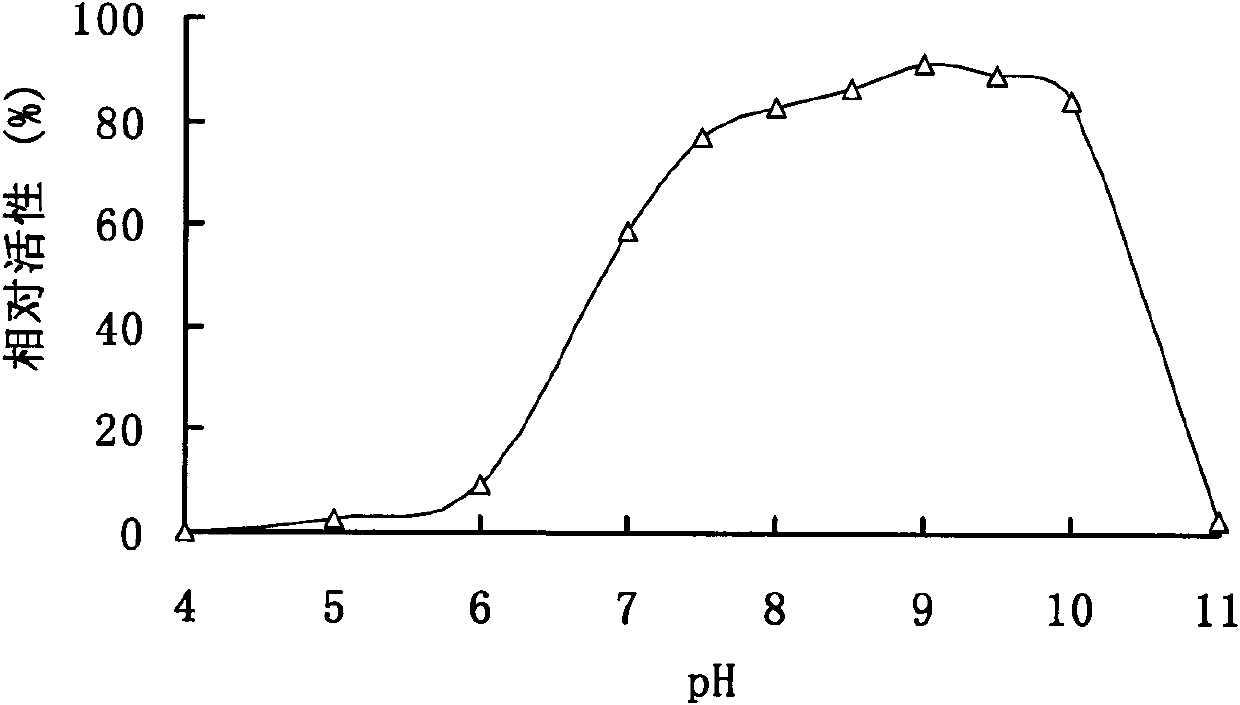
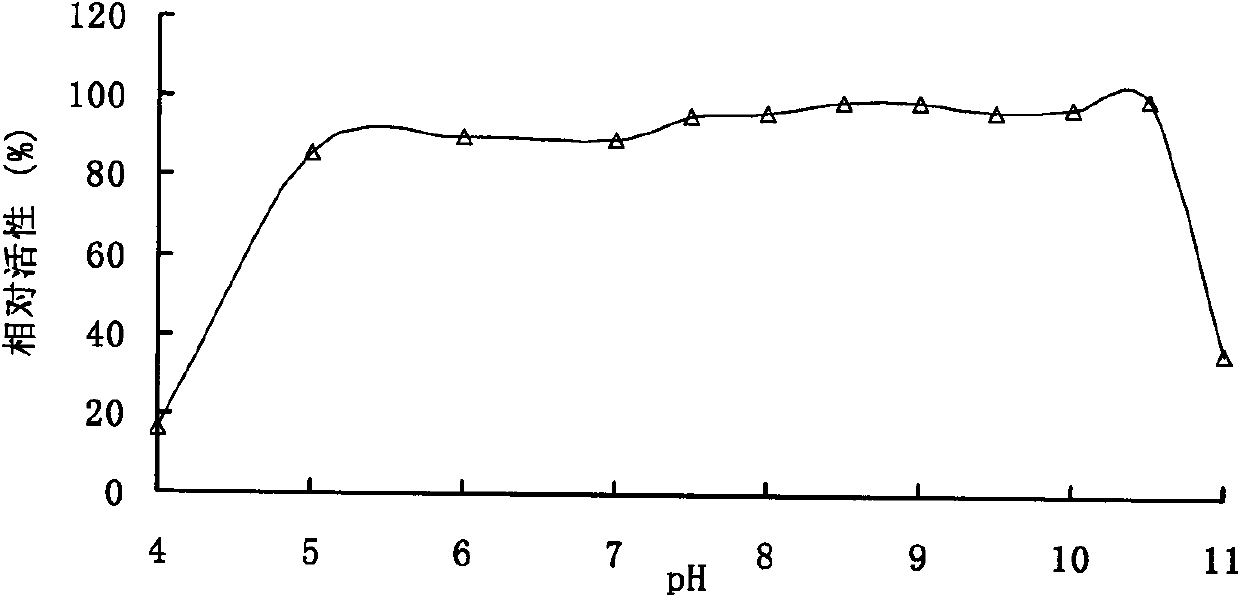
[0102] Enzyme activity analysis of embodiment 2 pectinase
[0103] The enzyme activity was determined by the DNS method. Under the conditions of pH 9.0 and 50°C, 1 mL of the reaction system includes 100 μL of appropriate diluted enzyme solution, 900 μL of substrate, reacted for 10 min, added 1.5 mL of DNS to terminate the reaction, and boiled for 5 min. After cooling, the OD value was measured at 540 nm. One enzyme activity unit (U) is defined as the amount of enzyme that releases 1 μmol of reducing sugar per minute under given conditions.
Embodiment 3
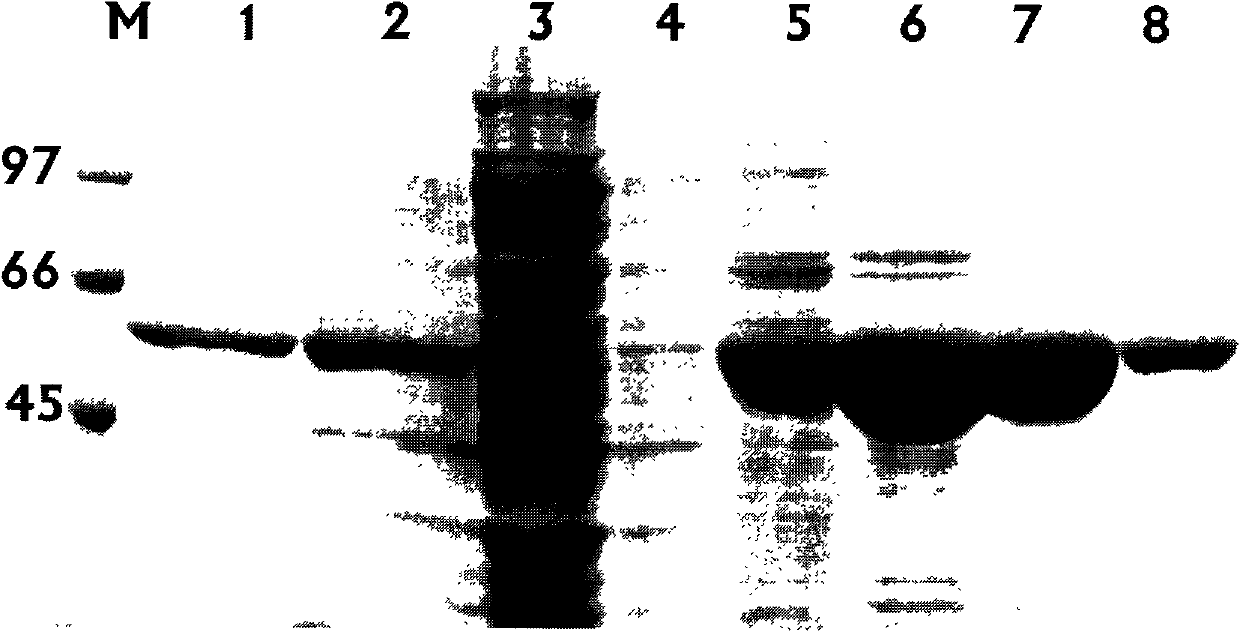
[0104] Example 3 Preparation of recombinant pectinase.
[0105] Design primers based on sequencing results:
[0106] PLAF (5'-CTG CCATGG CAGGAGAACGAGCGCCTGAGCAGCGTAAAGC-3', the underline is the Nco I site) and PLAR (5'-CTT GCGGCCGC TTATTTTTTGCCGTTCGGTTTCAGCTGCTCAC-3'), the underline is the Not I site) for PCR amplification. Plasmid pET22b(+) and gene fragments were digested and ligated to form vector pET22b(+)-plA to prepare BL21(DE3) competent cells, and the recombinant plasmid pET22b-plA was electroporated to transform the host bacteria. Positive recombinants were identified and induced to detect enzyme activity. Take the BL21(DE3) strain containing the recombinant plasmid and the BL21(DE3) strain containing the pET22b(+) empty plasmid (as a control), and inoculate them in 3 mL LB (adding 100 μg / mL ampicillin) culture solution, 37 Cultivate overnight with rapid shaking. Add 100 μL of overnight culture solution to 10 mL of LB culture solution (1% inoculum size) contain...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com