Multiple touchdown PCR (polymerase chain reaction) detection kit of haemophilus influenzae
A technology of Haemophilus influenzae and detection kit, which is applied in the field of molecular diagnosis of Haemophilus influenzae infection, and achieves the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
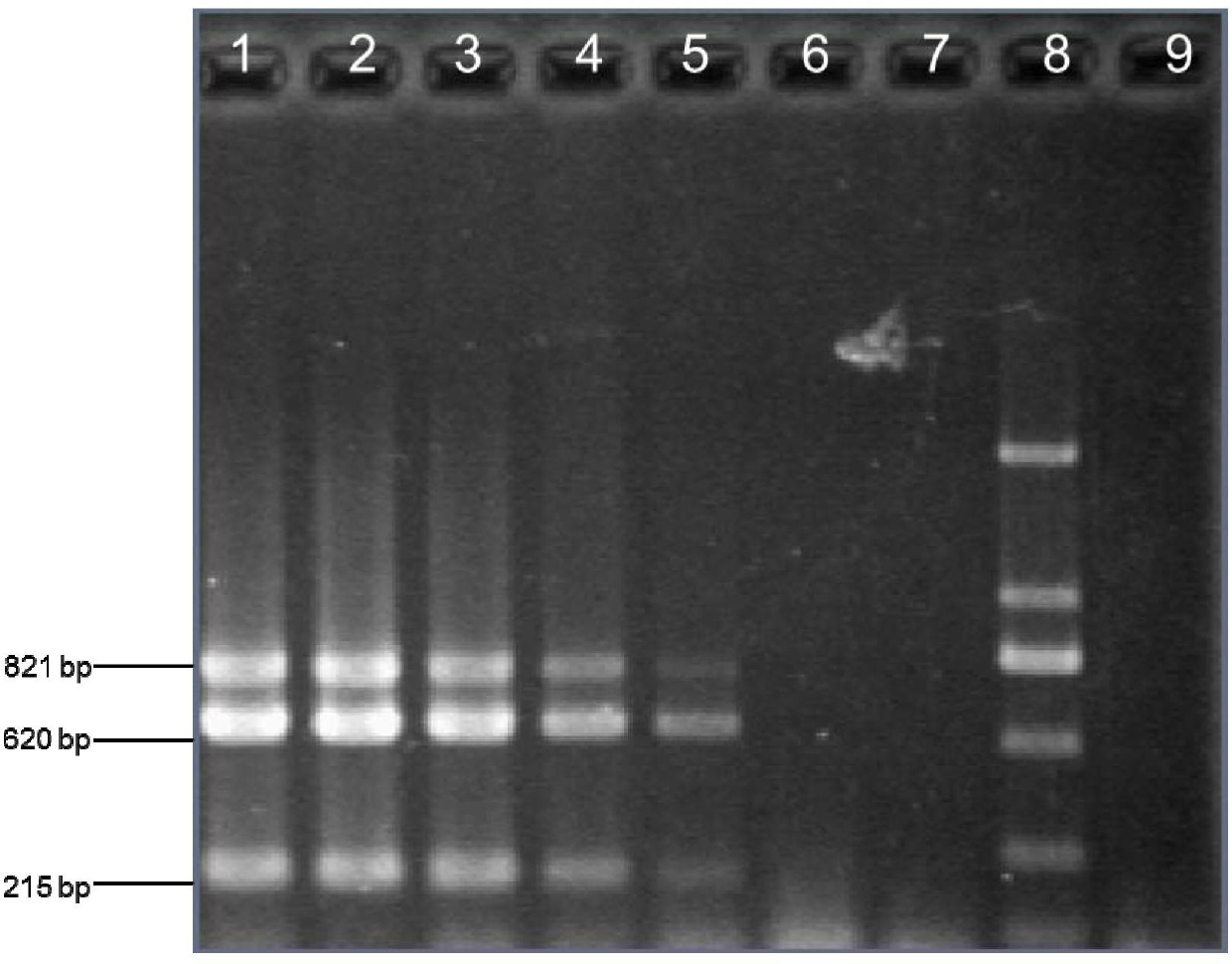
[0033] Pretreatment of sputum specimens: the sputum specimens should be washed twice with normal saline, added with an equal amount of 1% trypsin (prepared and used on the same day) and digested at 37°C for 90 minutes.
[0034] Bacterial DNA extraction from sputum samples: Take 1.5ml of digested sputum samples and use Takara minibest Bacterial Genome Extraction Kit (Takara, Baosheng, Dalian) to extract bacterial DNA from sputum samples. The extracted DNA was directly used for PCR amplification or stored at -20°C for future use.
[0035]PCR amplification total system 25μl, add 12.55μl double distilled water, 2.5μl buffer (10×PCR), BSA (10mg / ml) 0.25μl, primer Hi_omp P6-F (10μmol / L) 0.5 μl, Hi_omp P6-R (10 μmol / L) 0.5 μl, Hi_frdB-F (10 μmol / L) 1 μl, Hi_frdB-R (10 μmol / L) 1 μl, Hi_16S rRNA-F (10 μmol / L) 1 μl, Hi_16S rRNA-R ( 10μmol / L) 1μl, 2μl MgCl 2 (25mM), 0.5μl dNTP (10mM), 0.2μl Taq DNA polymerase (5U / μl), 2μl genomic DNA extracted from sputum samples. At the same time, a ...
Embodiment 2
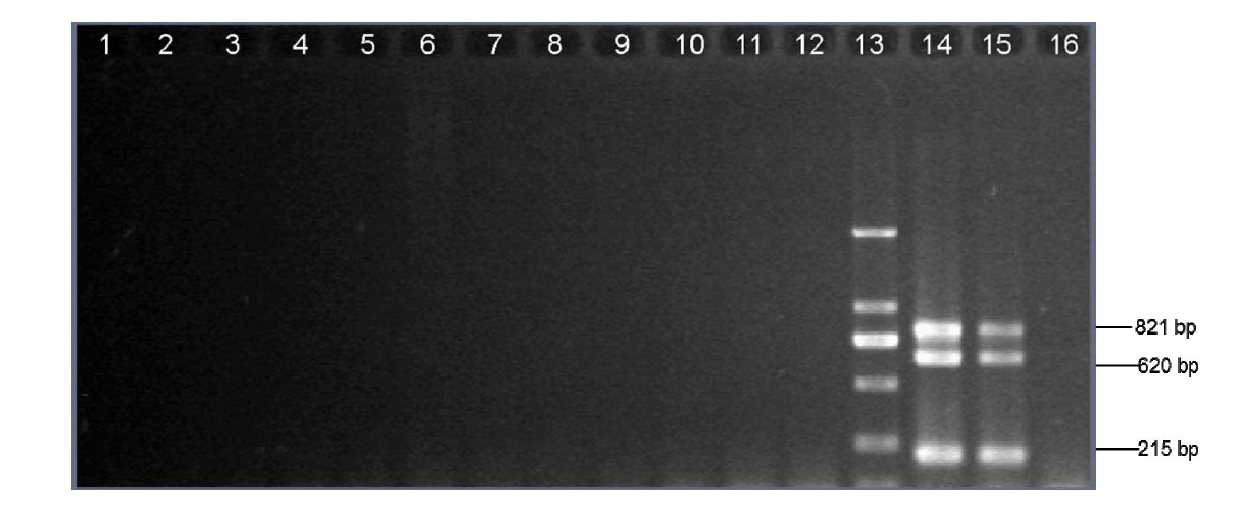
[0043] Specific detection by multiplex touchdown PCR.
[0044] Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, Klebsiella pneumoniae, Acinetobacter baumannii, Moraxella catarrhalis, Enterococcus faecalis, Mycobacterium smegmatis, Streptococcus pneumoniae, Genomic DNA of Mycobacterium tuberculosis, Mycobacterium bovis, Mycobacterium bovis BCG, Haemophilus influenzae and Haemophilus influenzae type b, the pure culture of the above bacteria was extracted with Takaraminibest Bacterial Genome Extraction Kit (Takara, Baosheng, Dalian) ) to extract its genomic DNA. The extracted DNA was directly used for PCR amplification or stored at -20°C for future use.
[0045] PCR amplification total system 25μl, add 12.55μl double distilled water, 2.5μl buffer (10×PCR), BSA (10mg / ml) 0.25μl, primer Hi_omp P6-F (10μmol / L) 0.5 μl, Hi_omp P6-R (10 μmol / L) 0.5 μl, Hi_frdB-F (10 μmol / L) 1 μl, Hi_frdB-R (10 μmol / L) 1 μl, Hi_16S rRNA-F (10 μmol / L) 1 μl, Hi_16S rRNA-R ( 10μmol / L) 1μ...
Embodiment 3
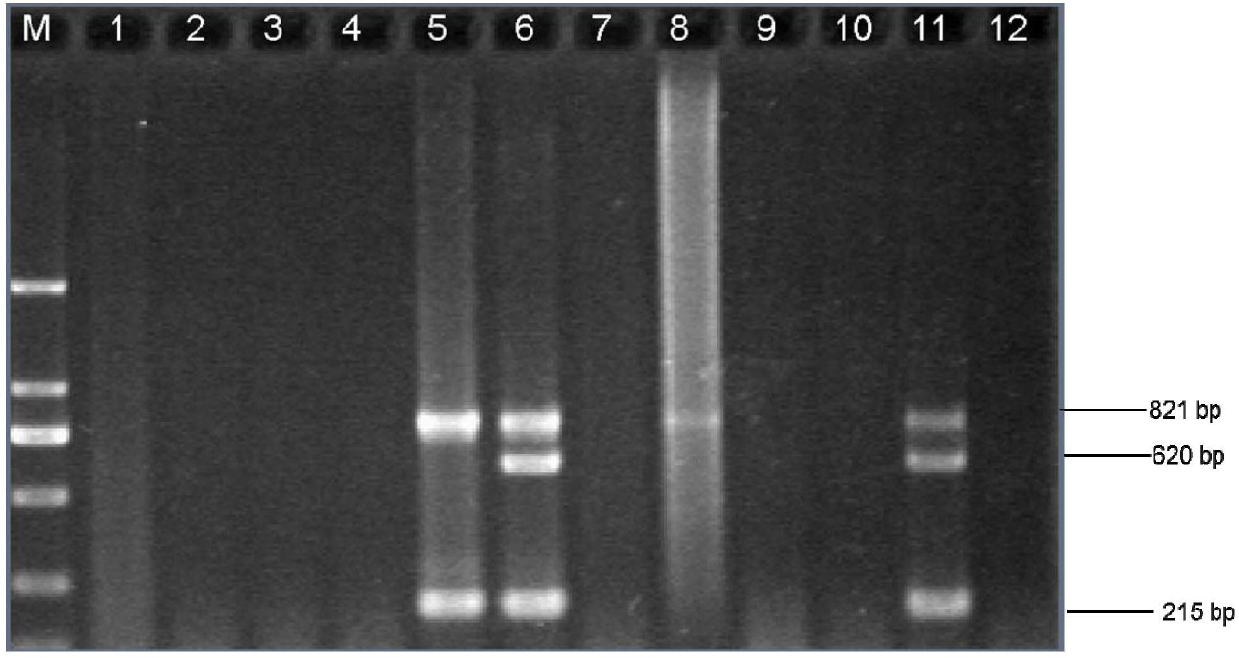
[0053] The detection limit of the multiplex touchdown PCR was determined.
[0054] Genomic DNA extraction of Haemophilus influenzae type b: Take 1.5 ml of pure culture solution of Haemophilus influenzae type b and use Takaraminibest Bacterial Genome Extraction Kit (Takara, Baosheng, Dalian) to extract its genomic DNA. The extracted DNA was directly used for PCR amplification or stored at -20°C for future use.
[0055] PCR amplification total system 25μl, add 12.55μl double distilled water, 2.5μl buffer (10×PCR), BSA (10mg / ml) 0.25μl, primer Hi_omp P6-F (10μmol / L) 0.5 μl, Hi_omp P6-R (10 μmol / L) 0.5 μl, Hi_frdB-F (10 μmol / L) 1 μl, Hi_frdB-R (10 μmol / L) 1 μl, Hi_16S rRNA-F (10 μmol / L) 1 μl, Hi_16S rRNA-R ( 10μmol / L) 1μl, 2μl MgCl 2 (25mM), 0.5μl dNTP (10mM), 0.2μl Taq DNA polymerase (5U / μl), the template is 2μl extracted Haemophilus influenzae type b genomic DNA or 10 -1~-6 Dilute the template. At the same time, a negative control (double distilled water as a template) was s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com