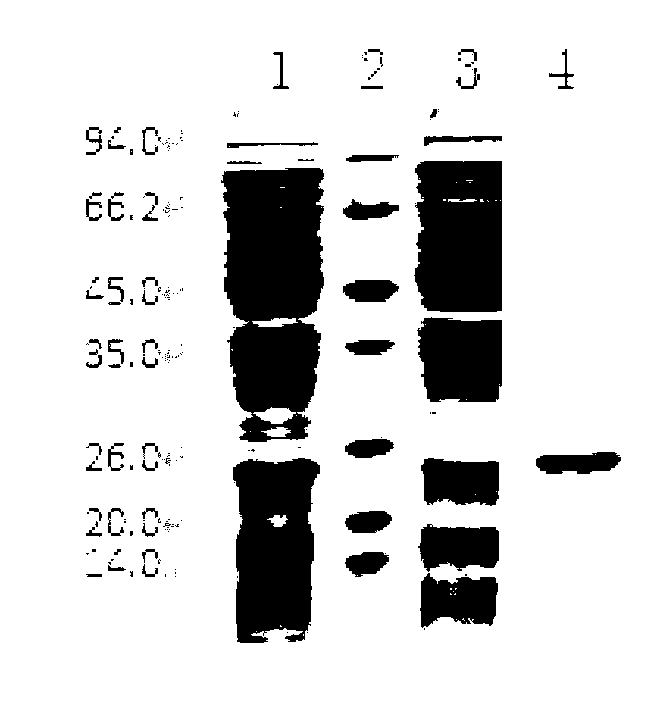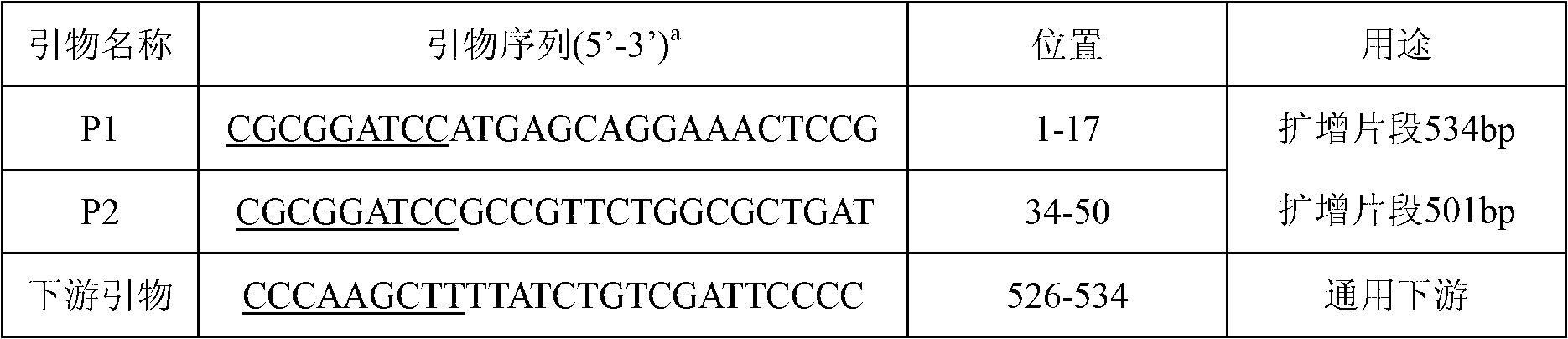Bacteria-cracking preparation for effectively cracking escherichia coli as well as cracking method and application thereof
A technology of Escherichia coli and bacteria preparations, which is applied in the field of preparation of bacteriostasis preparations, and can solve problems such as hindering the screening of bacteriostasis agents
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Embodiment 1, induction of phage and preparation of phage DNA
[0026] Through the method of phage induction, induce the lysogenic strain of Escherichia coli, obtain lytic phage particles, purify the phage, and extract the DNA of the phage; the specific operation is as follows:
[0027] Escherichia coli O157Min27 strain was inoculated into 5 mL of LB liquid medium, shaken overnight at 37°C, then added to fresh LB liquid medium at a volume ratio of 1:4, then added mitomycin C to a final concentration of 1.0 μg / mL, and continued shaking After shaking for 14 hours, add 0.1% chloroform, shake again for 15 minutes, centrifuge at 4000 g for 10 minutes, filter the supernatant to sterilize, and the filtrate contains Stx2 phage. Add MC1061 to LB medium for doubling dilution 10 -2 、10 -4 、10 -6 , take 200μl phage dilution and mix with 200μl host bacteria, and add 1mol / L CaCl 2 To a final concentration of 0.1mol / L, after incubating at 37°C for 15 minutes, add the mixture to ...
Embodiment 2
[0029] Embodiment 2, design 2 pairs of primer primers, PCR amplifies target gene fragment
[0030] Design 2 pairs of primers P1 and P2, use the DNA obtained in Example 1 as a template, apply PCR to amplify the corresponding gene fragments, and determine that the amplified fragments of the genes are 534bp and 501bp; the specific operations are as follows:
[0031] According to the DNA sequence of Escherichia coli O157Min27 phage in GenBank (NC_010237), 2 pairs of primers were designed. The primer sequences are shown in Table 1 to amplify 2 gene segments respectively. At the 34-534 amino acids of the position, BamH I and Hind III restriction sites were respectively introduced at the 5' ends of the upstream and downstream primers. Using phage DNA as a template, the target gene fragment was amplified according to conventional methods. After the reaction, 5 μL of the product was taken for agarose gel electrophoresis. PCR products were recovered using a PCR product recovery kit. ...
Embodiment 3
[0034] Embodiment 3, the construction of recombinant plasmid
[0035] Using the prokaryotic expression system pET-28a(+), the DNA amplified in Example 2 was used to construct recombinant plasmids, and sequence analysis and comparison were performed to determine the recombinant plasmids pET-28a(+)-P1 and pET-28a( +) P2; the specific operation is as follows:
[0036] The PCR amplification product of Example 2 was connected to the pMD-18T vector (Takara Company) for sequencing. The designed primers include a BamH I restriction site upstream and a Hind III restriction site downstream. The PCR product digested with BamHI and Hind III was ligated with pET-28a (+), T4 DNA ligase (Takara Company) overnight at 10-16°C. The ligated product was transformed into Escherichia coli DH5α, cultivated overnight at 37°C, and the plasmid was extracted. The identified positive recombinant plasmids were sequenced by digestion with BamH I and Hind III, and the positive recombinant plasmids pET-28...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com