Method and chip for detecting tumor cell EGFR (epidermal growth factor receptor) gene mutation
A tumor cell and chip technology, which is applied in biochemical equipment and methods, microbial determination/examination, chemical libraries, etc., can solve the problems of time-consuming, labor-intensive, high cost, and complicated operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Embodiment 1: chip preparation
[0065] Materials and reagents: Aldehyde glass substrate, gene microarray hybridization box, chip cover slip, chip fence, microarray chip centrifuge tube (purchased from Beijing Boao Biotechnology Company); chip spotting instrument (SpotBot Personal Microarray Robot) ) are products of Telechem Corporation.
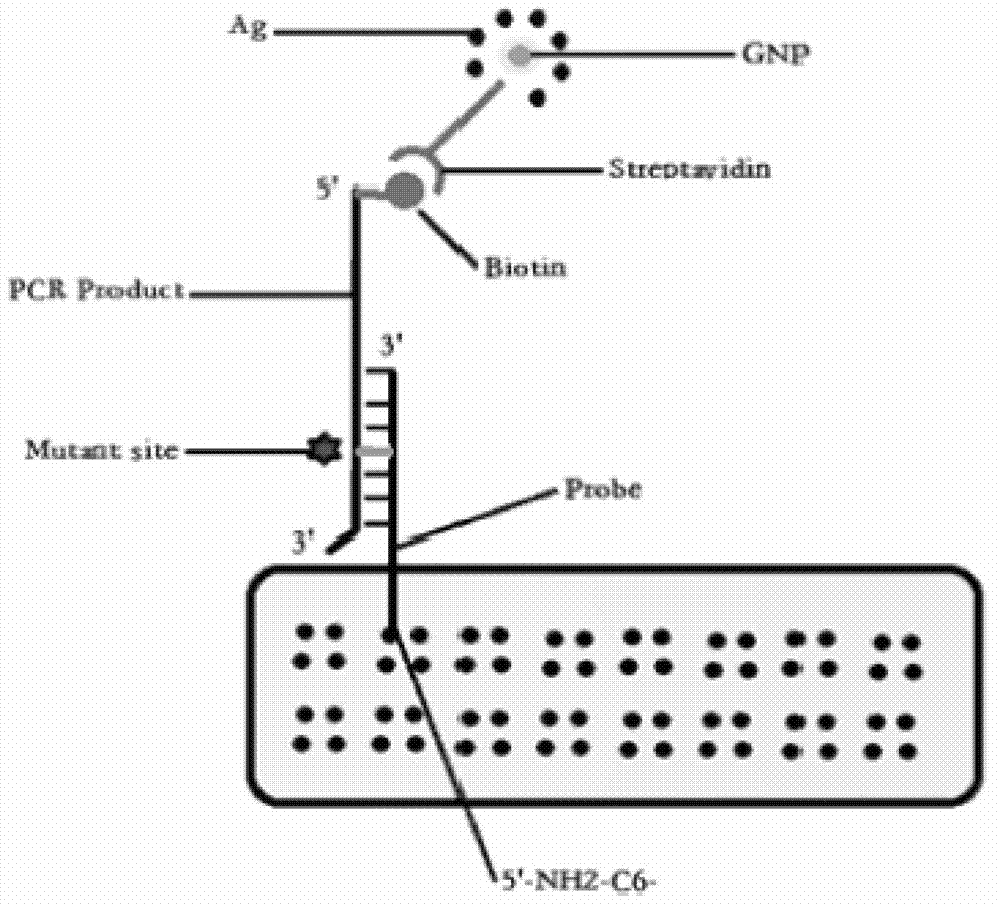
[0066] Oligonucleotide probe design and synthesis: use OLIGO6.0 software to design oligonucleotide probes (see SEQ ID NO:9-24), 5'-terminal amino modification (5'-NH 2 -(CH 2 ) 6 -), the specificity of all probes was confirmed by BLAST comparative analysis, and the probes were synthesized and purified by HPLC by Yingjun Biotechnology Company.
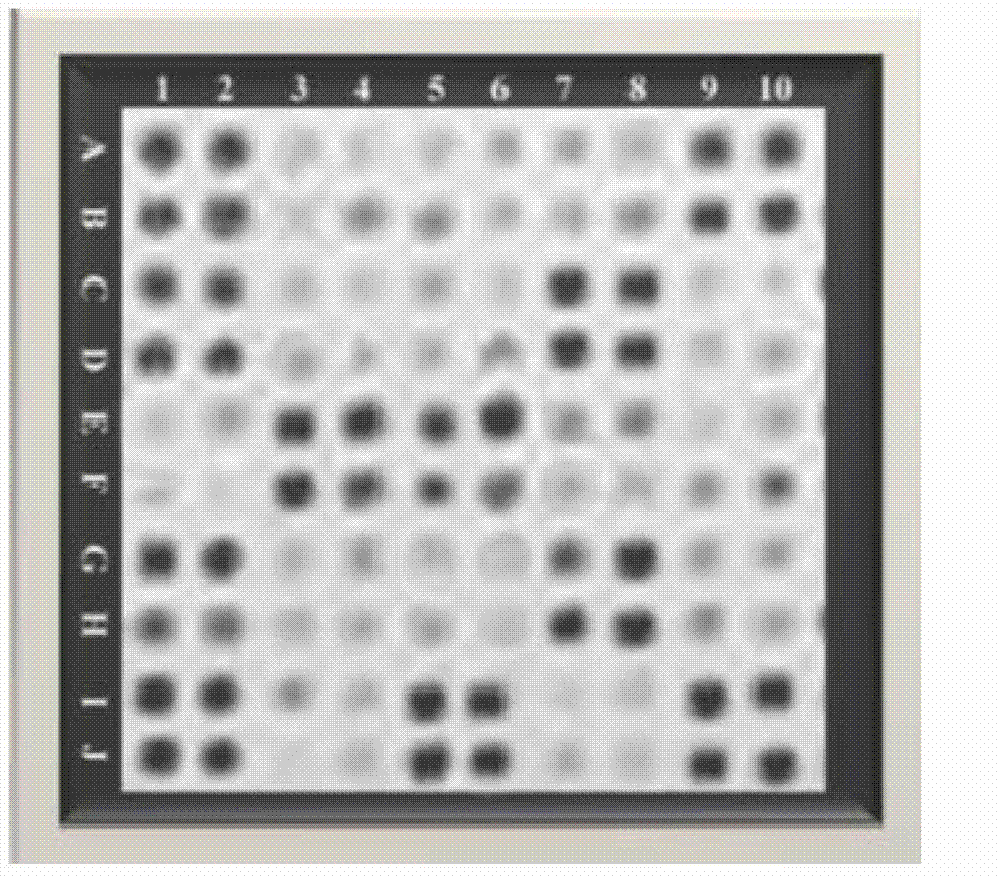
[0067] Chip preparation: 5'-NH 2 -(CH 2 ) 6 -Modify each probe, dilute to 20 μmol / L with spotting solution, use SpotBot Personal Microarray Robot automatic spotting instrument, SMP10B spotting needle to load the probe on the aldehyde glass slide, spotting diameter 365 μm, spacing 460 μm, ...
Embodiment 2
[0068] Embodiment 2: Tumor cell DNA extraction and PCR amplification
[0069] Sample to be tested: paraffin-embedded formalin-fixed (FFPE) tumor tissue;
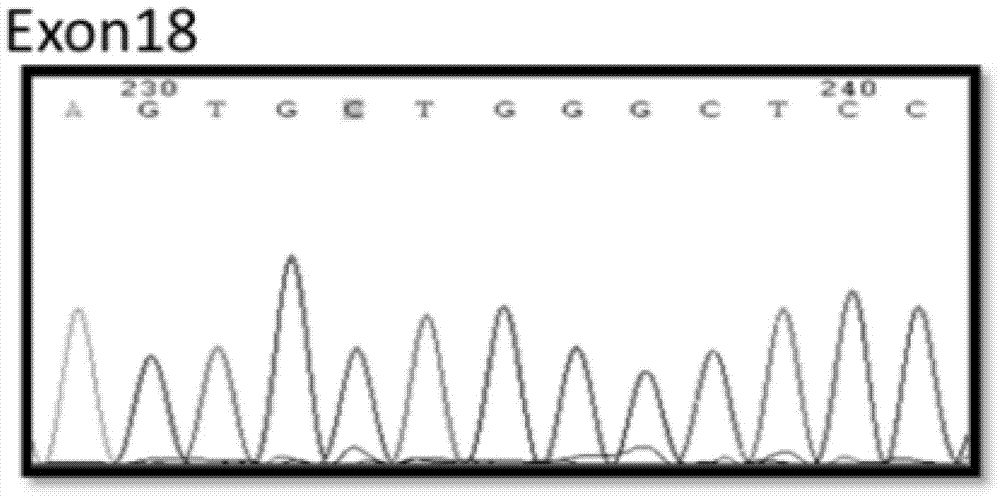
[0070] First deparaffinize, put 5-8 slices of 5-10μm thick FFPE tumor tissue into a 1.5ml centrifuge tube, incubate at 72°C for 20min, remove the dissolved paraffin liquid with a pipette, add 1ml xylene preheated to 50°C Solution, incubated at 50°C for 10 minutes, centrifuged at 12000g for 2 minutes, and then sucked off the xylene with a pipette. Repeat the operation of heating and removing the paraffin liquid 2 times. Add 1ml of absolute ethanol and place it at room temperature for 10min, centrifuge at 12000g for 5min, absorb the ethanol with a pipette, then add 90% and 70% ethanol in turn to repeat the hydration treatment, and finally wash and dry with deionized water. DNA was extracted with a paraffin-embedded tissue DNA extraction kit (QIAamp DNA FFPE tissue kit, QIAGEN) and operated according to the instructions. The...
Embodiment 3
[0072] Example 3: Preparation of streptavidin-coupled gold nanoparticles (Streptavidin-AuNP)
[0073] The pH value of the gold nanoparticle solution was adjusted to 6.5 with hydrochloric acid, and 100 μl (250 ng / μl) streptavidin was added to every 100 μl AuNP solution, and gently shaken at room temperature for 5 min. Add 100μl 10% NaCl solution and incubate at room temperature for 2h. Add 5% BSA to a final concentration of 2%, incubate at room temperature for 15 min, centrifuge at 20,000 rpm for 45 min at 4°C, and discard the supernatant. Sodium azide buffer (20mM Tirs-HCl pH6.2, 2%BSA, 0.02%NaN 3 ) to wash the red precipitate, centrifuge at 20,000 rpm for 45 min at 4°C, and discard the supernatant. Repeat washing and centrifugation 3 times, then resuspend the pellet with sodium azide buffer, and store at 4°C for future use.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com