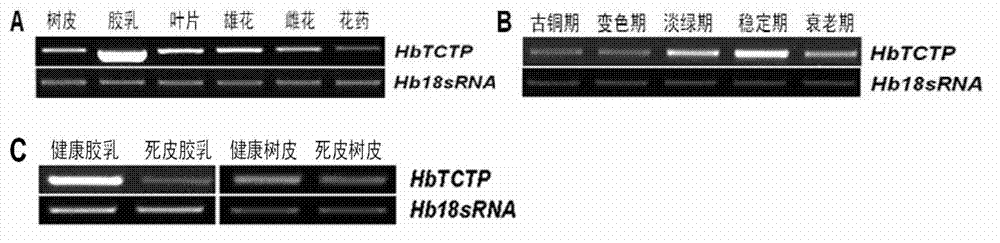
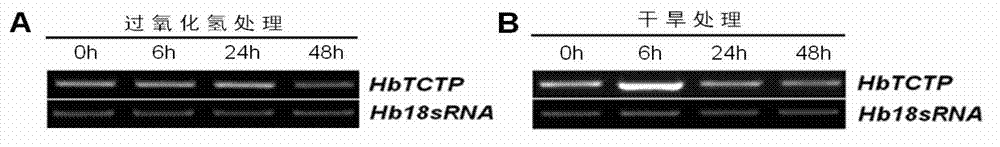
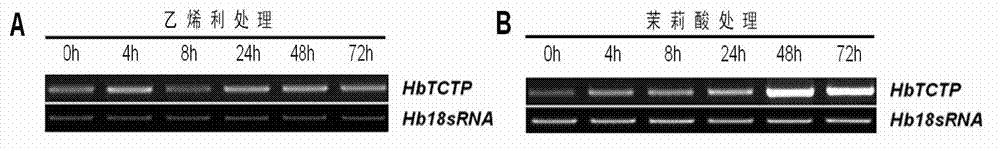
Rubber plant translationally controlled tumor protein, encoding gene thereof and application of rubber plant translationally controlled tumor protein
A protein and gene technology, applied in the rubber tree translation control tumor protein and its encoding gene and application field, can solve the problems of dead skin, latex coagulation, blocked milk ducts, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Embodiment 1, the acquisition of HbTCTP protein and its coding gene
[0040] 1. Acquisition of cDNA of HbTCTP
[0041] By searching the established Hevea bark transcriptome deep sequencing database, a 803bp EST fragment of the translational control tumor protein gene was found. DNAstar7.0 software analysis showed that the EST fragment contained a complete open reading frame. Using Hevea brasiliensis Reyan 7-33-97 (planted in the test field of Chinese Academy of Tropical Agricultural Sciences and the National Rubber Tree Germplasm Resource Garden) as the experimental material, total RNA was extracted from its bark and reverse transcribed into cDNA. The cDNA was used as a template, and 5'-tgttgctgcttcttctctcct-3' and 5'-agaacaaaacaacaactatataaac-3' were used as primers for PCR amplification. The reaction mixture is as follows:
[0042] wxya 2 o
18.3μl
10×PCR buffer
2.5μl
dNTP Mixture (2.5mM)
2.5μl
Taq enzyme (5U / μl)
0.2μl...
Embodiment 2
[0052] Embodiment 2, functional verification of HbTCTP
[0053] 1. Prokaryotic expression and purification of HbTCTP
[0054] Primers OTLF: 5'-ggatccatgttggtctatcaggatttgc-3' and OTFR: 5'-gaattcttagcatttgacctccttcaaag-3' were designed according to the HbTCTP open reading frame, and the cDNA of the bark of Hevea Reyan 7-33-97 was used as a template for PCR amplification with high-fidelity enzymes , to obtain a PCR product of about 500bp.
[0055] Connect the above PCR product to the pMD18-T vector, transform E.coli DH5α competent cells, spread on the LB plate containing ampicillin (100mg / L), obtain a band of about 500bp by colony PCR, and identify the colony by PCR The monoclonal extracted plasmid was double-digested with EcoR I and BamHI to obtain a band of about 500bp, and the colony PCR and enzyme digestion identified positive clones to the company for sequencing to obtain the recombinant plasmid pMD18-HbTCTP. Use EcoR I and RanH I to cut out the target fragment on the rec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com