Cephalosporin C acylase mutant as well as coding gene and application thereof
A technology of acylase mutants and cephalosporins, which is applied in the fields of application, genetic engineering, and plant gene improvement, and can solve problems such as incompetence for industrial production and low activity of cephalosporin acylases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] The acquisition of embodiment 1 cephalosporin C acylase gene
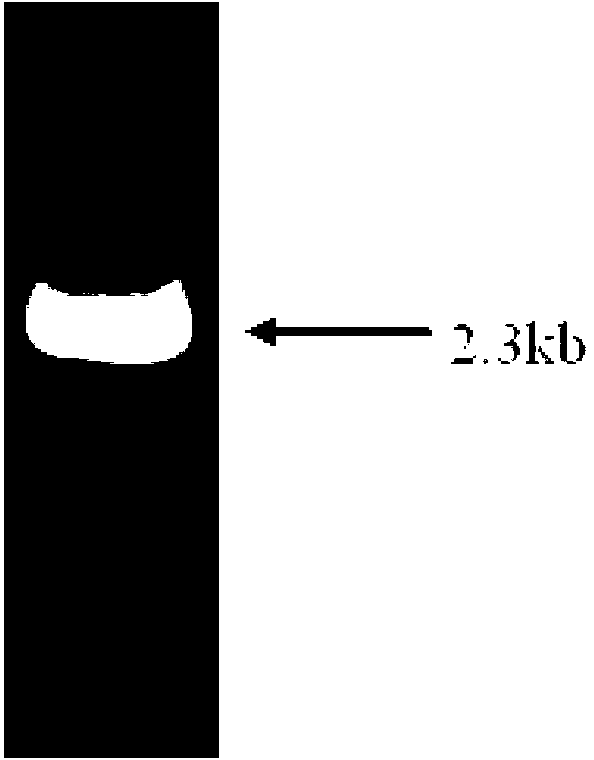
[0041] Using 1 μg of Pseudomonas sp. SE83 genomic DNA as a PCR reaction template, design the forward primer CPC-F as 5′-AAA according to the sequence of SEQ ID NO.1 ACGATGGCGGCCA-3'; reverse primer: 5'-AAA TCAATGATGATGATGATGATGATGGGCCGGGACGAGTTCCTGCGAG-3'. The parts in italics are the enzyme cutting sites NdeI and XhoI respectively. The PCR reaction was carried out in a total volume of 50 μL. The reaction conditions were 30 cycles of denaturation at 94°C for 5 min, denaturation at 94°C for 50 s, annealing at 58°C for 1 min, and extension at 72°C for 2 min. Take 3 μL of PCR amplification products for agarose gel electrophoresis verification, the results are as follows figure 1 shown. Take 100 μL of the PCR product for agarose gel electrophoresis, and recover the target fragment according to the steps of the gel recovery kit.
Embodiment 2
[0042] Embodiment 2 Construction of wild-type cephalosporin C acylase gene expression vector
[0043]After the PCR product in Example 1 was digested by restriction endonucleases NdeI and XhoI, it was ligated with the pET-30a plasmid (purchased from Novagen) digested with the same endonuclease, and the constructed vector was called pET -CPCA, then transform Escherichia coli DH5-a (purchased from Promega) with the ligation product pET-CPCA mixture, and perform sequence determination to extract the plasmid of the transformed cell library, and transform Escherichia coli BL (DE3) strain (purchased from Novagen) .
Embodiment 3
[0044] Example 3 Error-prone PCR amplification of cephalosporin C acylase gene
[0045] Using the property that TaqDNA polymerase does not have 3′-5′ proofreading function, under high magnesium ion concentration (5mmol / L-9mmol / L) and different concentrations of dNTP (1.5mmol / L-3.5mmol / L), to Control the frequency of random mutations, introduce random mutations into the target gene, and construct a mutation library. The optimal mutation rate in the experiment is about 0.6%.
[0046] Error-prone PCR reaction system (100μl):
[0047]
[0048]
[0049] The PCR program was: pre-denaturation at 96°C for 4 min, denaturation at 94°C for 1 min, annealing at 56°C for 1 min, extension at 75°C for 1 min and 40 s, 45 cycles of reaction, and finally extension at 75°C for 15 min. The PCR product was recovered by gel recovery method. Take 5 μl of the product for 1% agarose gel electrophoresis, and store it at -20°C for later use.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com