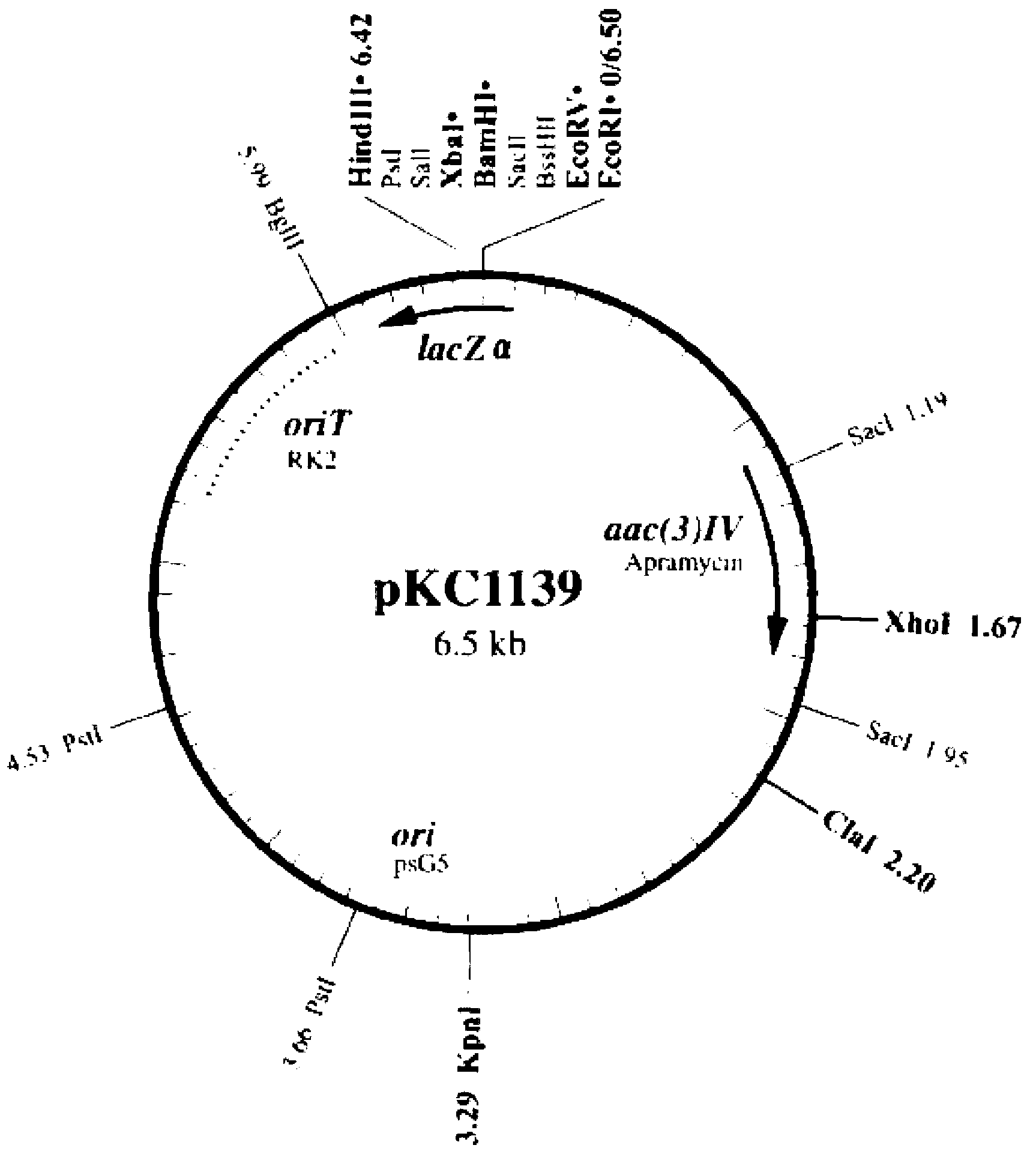
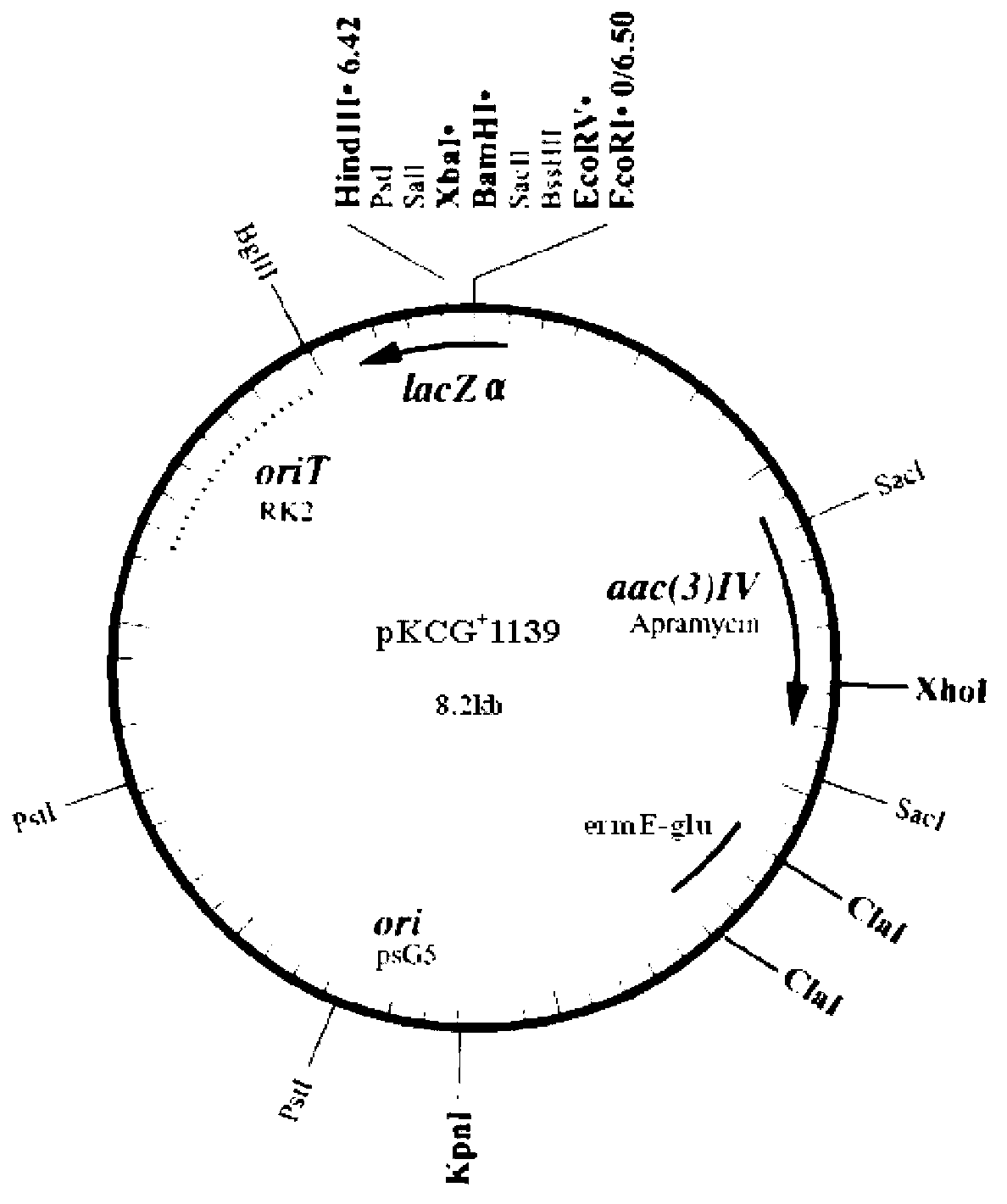
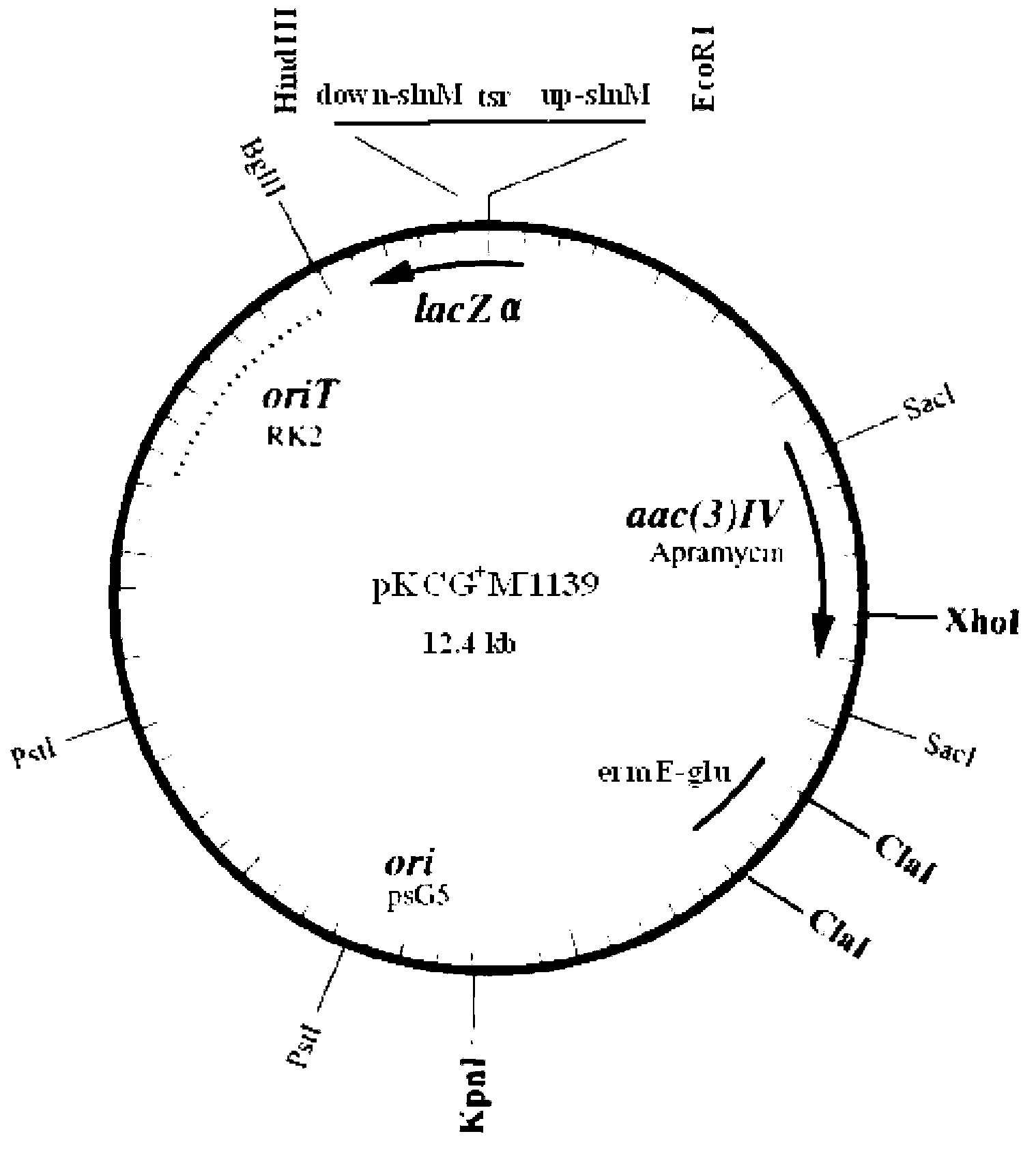
Vector for knocking out streptomycete gene as well as constructing method and application of same
A Streptomyces and gene technology, applied in the field of Streptomyces gene knockout vectors, can solve the problems of poor colony PCR stability, false positives, inability to improve the screening efficiency of double-crossover transformants, etc., to achieve simple gene knockout and make up for false positives Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Embodiment 1, construction is used for knocking out the knockout vector of the Streptomyces gene that does not produce glucanase
[0055] 1. Cloning of Bacillus amyloliquefaciens glucanase gene
[0056] Using a Bacillus amyloliquefaciens YW26 genomic DNA screened by our laboratory that has significant inhibitory effects on a variety of plant pathogenic fungi as a template, according to the sequence of the Bacillus amyloliquefaciens endoglucanase gene (DQ782954.1) in GenBank Design primers: P1 (5'-ATGAAACGGTCAATTTCTAT-3') and P2 (5'-CTAATTGGGTTCTGATCC-3') for PCR amplification. The reaction was pre-denatured at 95°C for 5min; 30 cycles (94°C for 40s, 55°C for 30s, 72°C for 1min); 72°C for 10min. The amplified 1500bp fragment was ligated with the plasmid pMD18-T vector to obtain pMD18-glu, which was transformed into Escherichia coli DH5α, and the obtained positive transformants were verified by PCR and enzyme digestion through ampicillin resistance screening. Sequencing...
Embodiment 2
[0062] Example 2. Construction of a knockout vector for knockout of the positive regulatory gene slnM for natamycin synthesis in Streptomyces that does not produce glucanase
[0063] The coding sequence of the natamycin synthesis positive regulatory gene slnM is SEQ ID No.4, which encodes a protein whose amino acid sequence is shown in SEQ ID No.3.
[0064] Firstly, the nucleotide sequence is constructed as a DNA fragment of the knockout Streptomyces slnM of SEQ ID No.5. SEQ ID No.5 consists of 4144 nucleotides, and positions 1-8 are EcoRI recognition sites and protected bases. Positions 29-1221 are the upstream homology arm of the natamycin synthesis positive regulatory gene slnM, positions 1265-2911 are thiostrepton resistance gene (tsr), and positions 2956-4117 are positive regulators of natamycin synthesis In the downstream homology arm of the regulatory gene slnM, positions 4136-4144 are HindIII recognition sites and protective bases.
[0065] The DNA fragment of the kno...
Embodiment 3
[0066] Example 3. Construction of a mutant strain in which the positive regulator gene slnM for natamycin synthesis in streptomyces that does not produce glucanase is knocked out
[0067] 1. The pKCG + m - 1139 vector transformed demethylated E.coliET12567 (pUZ8002)
[0068] pKCG by heat shock method + m - The 1139 vector was introduced into E.coliET12567 (pUZ8002) and was screened for resistance to 15 μg / ml thiostrepton to obtain pKCG + m - 1139 vector recombinant strain E.coliET12567(pUZ8002) / pKCG + m - 1139.
[0069] 2. Preparation of slnM knockout strain
[0070] Reference (Bierman M et al.1992) for amphiphilic conjugation pKCG + m - 1139 was introduced into Streptomyces lydidii, and the specific method was as follows:
[0071] The recombinant strain E.coliET12567(pUZ8002) / pKCG + m - 1139 was inoculated into LB liquid medium containing chloramphenicol, kanamycin and apramycin resistance, cultured overnight at 37°C, 200rpm, and transferred to fresh LB medium at...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com