Probe, primer and detection kit for detecting C-KIT gene mutation
A detection kit, the technology of the kit, applied in the fields of DNA/RNA fragments, fluorescence/phosphorescence, recombinant DNA technology, etc., can solve the problems of long detection time, false negative, inability to meet, etc., and achieve fast detection speed and strong specificity. , the effect of high stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
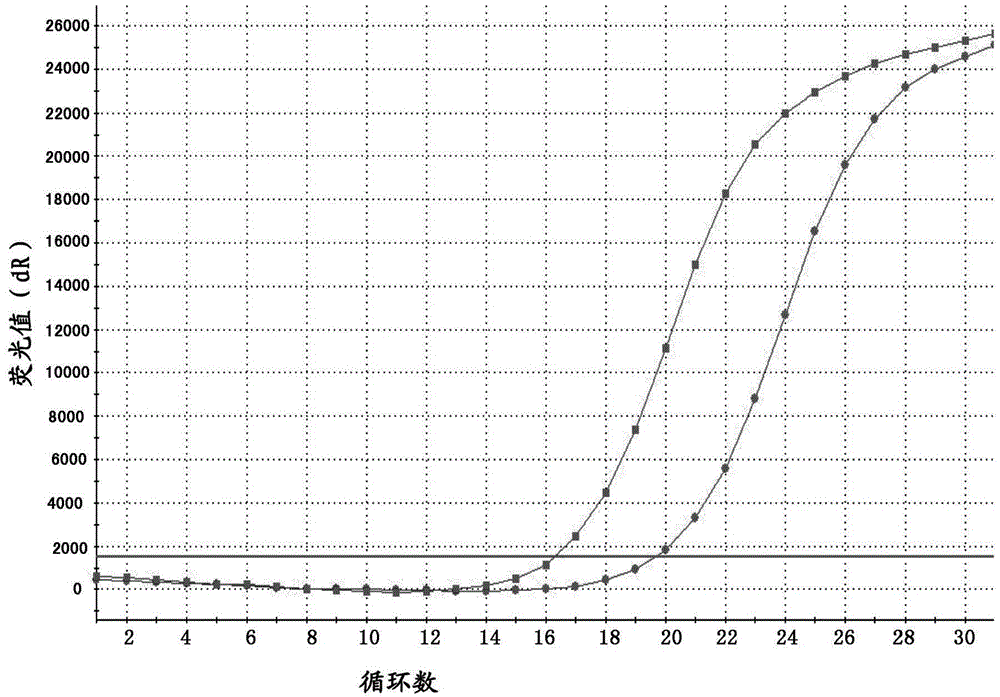
[0037] In this embodiment, the plasmid constructed by genetic engineering is used as the positive plasmid, and the method for detecting the D842V mutation of the C-KIT gene by real-time fluorescent PCR of the present invention includes the following steps:
[0038] (1) Detection sample DNA extraction
[0039] Add 1 mL of xylene to the sample for dewaxing, centrifuge to collect the precipitate, add 1 mL of absolute ethanol to the precipitate, dry at room temperature or 37°C, add proteinase K and Buffer ATL, digest and lyse at 56°C for 1 hour, incubate at 90°C for 1 hour, add 200mL Mix Buffer AL, then add 200 μL absolute ethanol and mix well, carefully transfer the supernatant to a QIA 2mL spin column, centrifuge at 6000×g (8000rpm) for 1min, add 500μL BufferAW1, centrifuge at 6000×g (8000rpm) for 1min, carefully open the lid and add Centrifuge 500μL Buffer AW2 at 6000×g (8000rpm) for 1min, centrifuge the empty tube at 20000×g (14000rpm) for 3min, add 100μL Buffer ATE to the cen...
Embodiment 2
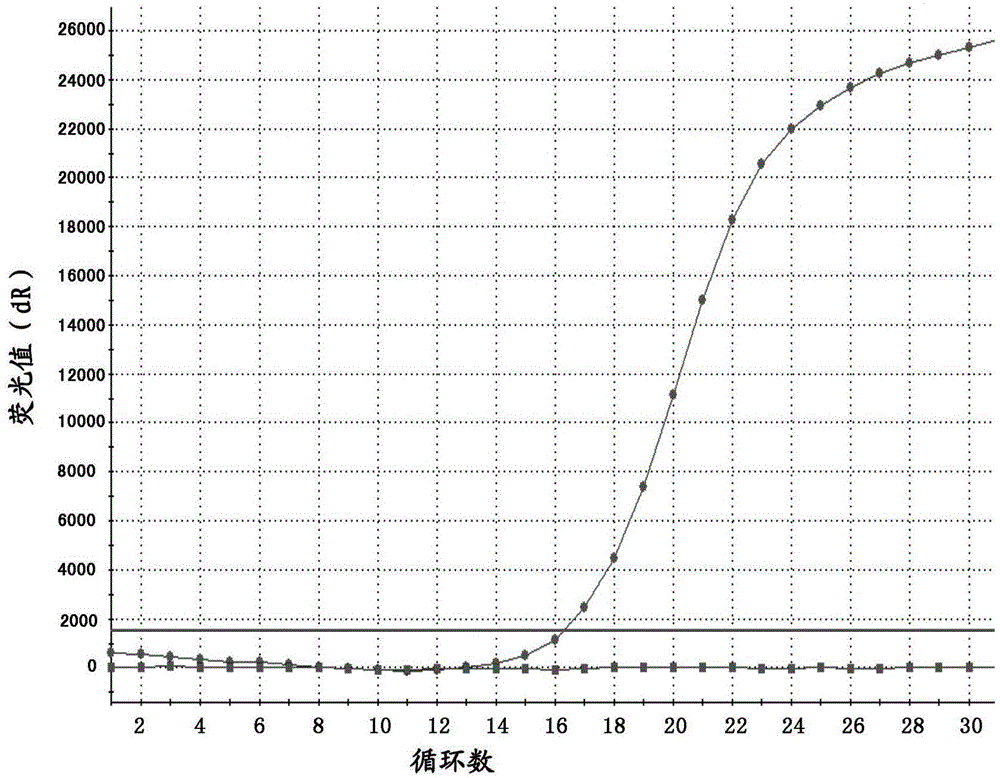
[0051] Using the present invention to detect clinical samples, a total of 90 clinical gastrointestinal stromal tumor samples from our company were taken, including 40 males and 50 females, aged 30-72 years old, with an average age of 50 years old. The steps of detecting C-KIT gene mutations in 90 cases of clinical samples by using the real-time fluorescent PCR system of the present invention are as follows.
[0052] (1) Detection sample DNA extraction
[0053] Test samples included fresh pathological tissue, paraffin-embedded tissue, whole blood, plasma, and pleural effusion. For fresh pathological tissue and paraffin tissue samples, cut the sample into 5-10 μm, add 1 mL of xylene to dewax, collect the precipitate by centrifugation, add 1 mL of absolute ethanol to the precipitate, dry at room temperature or 37°C, add proteinase K and BufferATL, 56 Digest and lyse at ℃ for 1 hour, incubate at 90°C for 1 hour, add 200 mL of Buffer AL to mix, then add 200 μL of absolute ethanol ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com